Practical 11¶
In this practical we will will see some other functionalities of Biopython.
Biopython¶
From the Biopython tutorial: The Biopython Project is an international association of developers of freely available Python tools for computational molecular biology. The goal of Biopython is to make it as easy as possible to use Python for bioinformatics by creating high-quality, reusable modules and classes. Biopython features include parsers for various Bioinformatics file formats (BLAST, Clustalw, FASTA, Genbank,…), access to online services (NCBI, Expasy,…), interfaces to common and not-so-common programs (Clustalw, DSSP, MSMS…), a standard sequence class, various clustering modules, a KD tree data structure etc. and even documentation.
In this practical we will see some features of Biopython but refer to biopython documentation to discover all its features, recipes etc.
These notes are largely based on what is available here.
BLAST¶
Blast (Basic logical alignment search tool) is a well known tool to find similarities between biological sequences. It compares DNA or protein sequences and calculates the statistical significance of the matches found.
The typical interaction with BLAST sees the user submit some sequences to the tool to get an alignment and then the hits are parsed to obtain information on the matches. Both these steps can be performed from within Biopython. Although it is possible to interact directly with a local installation of BLAST, in this practical we will work with the tool made available by NCBI (available here).
The function qblast¶
The online version of blast can be accessed through the
Bio.Blast.NCBIWWW.qblast() function.
It’s basic syntax is the following (first import
from Bio.Blast import NCBIWWW):
result_handle = Bio.Blast.NCBIWWW.qblast(blast_program, database, query_str)
where blast_program is the program to perform the alignment. The
options are blastn, blastp, blastx, tblast or tblastx. database
is the database to search against and query_str is a string
containing the query to search against the database. The query can be a
sequence or a fasta file entry or an identifier like a GI number
(NCIBI’s sequence identification number). Among the others, some
optional parameters are the output format (format_type that by
default is “XML” which is the most stable output format but results can
be stored also as text with “Text”). It is also possible to specify an
expectation value cut-off to filter out alignments expect (the
e-value threshold, default value is 10.0).
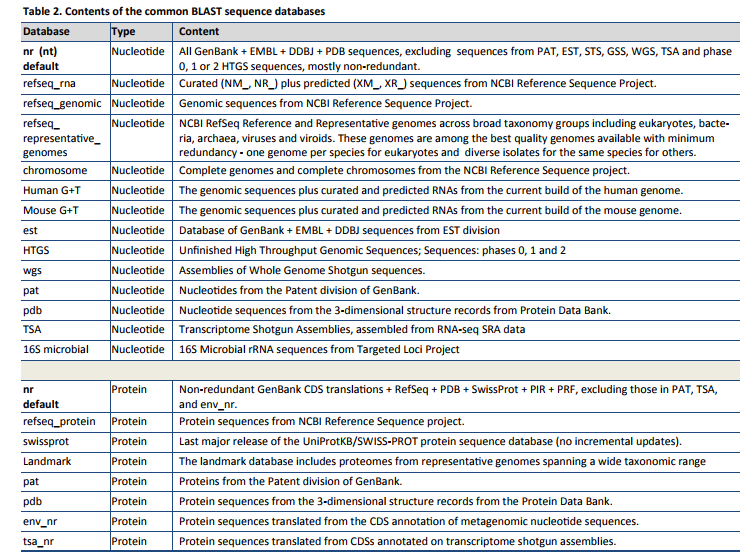
Some databases to search against are reported below:
The query string can be obtained in different ways, for example it is possible to load sequences from a fasta file with:
from Bio.Blast import NCBIWWW
fasta_string = open("myfile.fasta").read()
result_handle = NCBIWWW.qblast("blastn", "nt", fasta_string)
or we can give a SeqRecord:
from Bio.Blast import NCBIWWW
from Bio import SeqIO
record = SeqIO.read("myfile.fasta", format="fasta")
result_handle = NCBIWWW.qblast("blastn", "nt", record.seq)
It is also possible to specify some optional parameters in the
entrez_query for example we can limit the search to specific
organisms with: entrez_query='"Malus Domestica" [Organism]'.
Example: Let’s align the first 100 bases of the first entry of the file contigs82.fasta to the Malus Domestica genome.
NOTE: this can take several minutes.
In [2]:
from Bio.Blast import NCBIWWW
from Bio import SeqIO
records = SeqIO.parse("file_samples/contigs82.fasta", format="fasta")
rec = next(records)
seq = rec.seq[0:100]
print("Aligning {} [{}] to Malus Dom.".format(rec.id,
seq[0:10]+"..."+seq[90:101]))
result_handle = NCBIWWW.qblast("blastn", "nt",
seq,
entrez_query='"Malus Domestica" [Organism]'
)
Aligning MDC020656.85 [GAGGGGTTTA...TTGGCAGCAA] to Malus Dom.
Note that the previous code does not output anything, it just returns a
result_handle. We need to parse it to get some results.
Parsing qblast output¶
Once the qblast call returns, it gives the results in a handle object
result_handle that we can parse or we can write to disk to avoid
having to rerun the query other times. If we expect to get one alignment
only, we can use the method read otherwise (if we have multiple
query sequences) we should use the method parse:
blast_record = NCBIXML.read(result_handle)
or
blast_records = NCBIXML.parse(result_handle)
Note that to use these methods we first need to import the NCBIXML
module with from Bio.Blast import NCBIXML.
These methods are analogous to what seen in the case of SeqIO and AlignIO. In the case of multiple entries we can loop through them with:
blast_records = NCBIXML.parse(result_handle)
for record in blast_records:
#do something with it...
or we can retrieve one record at a time with
record = next(blast_records).
Saving results to file¶
To save the results present in the result_handle we can simply write them to file. In case we have only one entry we can read it and write it to file:
out_f = open("my_blast_result.xml", "w")
out_f.write(result_handle.read())
out_f.close
result_handle.close()
If we have more than one entry we need to loop through all the entries and save them in the file:
out_f = open("my_blast_result.xml", "w")
for entry in result_handle.parse():
out_f.write(entry)
out_f.close
result_handle.close()
Example:
Let’s BLAST the galactosidase alpha (gi number: 2717) against the human database on NCBI and save the results to file. (Note that it can take several seconds/minutes to run!).
In [38]:
from Bio.Blast import NCBIWWW
result_handle = NCBIWWW.qblast("blastn", "nt", "2717")
with open("file_samples/blast_res.xml","w") as out_f:
out_f.write(result_handle.read())
result_handle.close()
Open a blast .XML file¶
A BLAST output file can be read by opening the file to get the handler and then parse it with the method parse seen above:
from Bio.Blast import NCBIXML
result_handle = open("my_blast.xml")
blast_records = NCBIXML.parse(result_handle)
This will end up in a handle to the blast results.
The BLAST record class¶
The Bio.Blast.Record.Blast class holds the results of the alignment.
In particular it is composed of the following three information:
- query: the identifier of the query (a string).
- Descriptions : a list of Description objects. Each
Descriptionholds the following information:Description.title: a string with the title of the hit;Description.score: a float with the score of the alignment;Description.num_alignments: an int with the number of alignments with the same subject;Description.e: a float with the e-value of the alignment.
- Alignments : a list of Alignment objects. Each
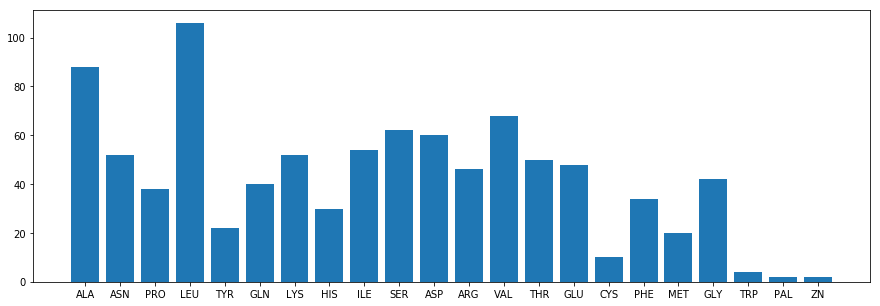
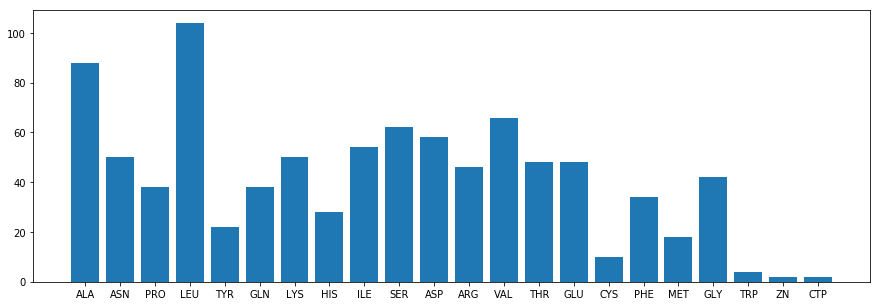
Alignmentholds the following information:Alignment.title: a string with the title of the hit (identical toDescription.title);Alignment.length: an int with the length of the alignment;Alignment.hsps: a list of HSP objects (High Scoring Pair). EachHSPhas the following info:HSP.score: the BLAST score of the hitHSP.bits: the bits score of the hit (x: on average 2^x pairs to find such a good hit by chance)HSP.expect: the evalue of the hitHSP.num_alignments: the number of alignments for the same subjectHSP.identities: the number of identities between query and subjectHSP.positives: the number of identical bases/aminos or having similar chemical propertiesHSP.gaps: the number of gaps between query and subjectHSP.strand: a tuple with (query,subject) strandsHSP.frame: a tuple with the frame shiftsHSP.query/HSP.sbjct: query/subject sequenceHSP.query_start/HSP.sbjct_start:query/subject start pointHSP.match: the match sequence (basically “|” for matches and spaces for mismatches)HSP.align_length: the alignment length.
More information on the BLAST record can be found here.
Example:
Let’s blast the serum albumin sequence (gi number 23307792) on the human genome and report all the information reported by BLAST. (warning might take a while to run!)
In [39]:
from Bio.Blast import NCBIWWW
from Bio.Blast import NCBIXML
result_handle = NCBIWWW.qblast("blastn", "nt", "23307792",
entrez_query='"Homo Sapiens" [Organism]'
)
for res in NCBIXML.parse(result_handle):
for d in res.descriptions:
print("TITLE:{}\nSCORE:{}\nN.ALIGN:{}\nE-VAL:{}".format(
d.title,d.score, d.num_alignments,d.e))
for a in res.alignments:
print("Align Title:{}\nAlign Len: {}".format(a.title, a.length))
for h in a.hsps:
s = h.score
b = h.bits
e = h.expect
n = h.num_alignments
i = h.identities
p = h.positives
g = h.gaps
st = h.strand
f = h.frame
q = h.query
sb = h.sbjct
qs = h.query_start
ss = h.sbjct_start
qe = h.query_end
se = h.sbjct_end
m = h.match
al = h.align_length
print("Score: {} Bits: {} E-val: {}".format(s,b,e))
print("N.aligns:{} Ident:{} Pos.:{} Gaps:{} Align len:{}".format(
n,i,p,g,al))
print("Strand: {} Frame: {}".format(st,f))
print("Query:", q, " start:", qs, " end:", qe)
print("Match:",m)
print("Subjc:",sb, " start:", ss, " end:", se)
result_handle.close()
TITLE:gi|23307792|gb|AF542069.1| Homo sapiens serum albumin (HSA) mRNA, complete cds
SCORE:4352.0
N.ALIGN:1
E-VAL:0.0
TITLE:gi|1519245814|ref|NM_000477.7| Homo sapiens albumin (ALB), mRNA
SCORE:4305.0
N.ALIGN:1
E-VAL:0.0
TITLE:gi|28591|emb|V00495.1| H.sapiens mRNA for serum albumin
SCORE:4253.0
N.ALIGN:2
E-VAL:0.0
TITLE:gi|7770116|gb|AF119840.1|AF119840 Homo sapiens PRO0903 mRNA, complete cds
SCORE:4062.0
N.ALIGN:1
E-VAL:0.0
TITLE:gi|25058738|gb|BC039235.1| Homo sapiens albumin, mRNA (cDNA clone IMAGE:4768004), containing frame-shift errors
SCORE:4056.0
N.ALIGN:1
E-VAL:0.0
TITLE:gi|23243417|gb|BC036003.1| Homo sapiens albumin, mRNA (cDNA clone MGC:32850 IMAGE:4724105), complete cds
SCORE:4052.0
N.ALIGN:1
E-VAL:0.0
TITLE:gi|158258946|dbj|AK292755.1| Homo sapiens cDNA FLJ78413 complete cds, highly similar to Homo sapiens albumin, mRNA
SCORE:4036.0
N.ALIGN:1
E-VAL:0.0
TITLE:gi|28589|emb|V00494.1| Human messenger RNA for serum albumin (HSA)
SCORE:4033.0
N.ALIGN:1
E-VAL:0.0
TITLE:gi|21706455|gb|BC034023.1| Homo sapiens albumin, mRNA (cDNA clone MGC:22784 IMAGE:4734617), complete cds
SCORE:4029.0
N.ALIGN:1
E-VAL:0.0
TITLE:gi|51476389|emb|CR749331.1| Homo sapiens mRNA; cDNA DKFZp779N1935 (from clone DKFZp779N1935)
SCORE:4027.0
N.ALIGN:1
E-VAL:0.0
TITLE:gi|21706470|gb|BC034026.1| Homo sapiens cDNA clone IMAGE:4734794, containing frame-shift errors
SCORE:4010.0
N.ALIGN:1
E-VAL:0.0
TITLE:gi|194391079|dbj|AK298437.1| Homo sapiens cDNA FLJ54371 complete cds, highly similar to Serum albumin precursor
SCORE:3994.0
N.ALIGN:1
E-VAL:0.0
TITLE:gi|408407943|gb|AF130077.2| Homo sapiens clone FLB9714 PRO2619 mRNA, complete cds
SCORE:3937.0
N.ALIGN:1
E-VAL:0.0
TITLE:gi|62113340|gb|AY960291.1| Homo sapiens serum albumin mRNA, complete cds
SCORE:3907.0
N.ALIGN:1
E-VAL:0.0
TITLE:gi|49176516|gb|AY550967.1| Homo sapiens cell growth inhibiting protein 42 mRNA, complete cds
SCORE:3848.0
N.ALIGN:1
E-VAL:0.0
TITLE:gi|115607206|gb|DQ986150.1| Homo sapiens serum albumin mRNA, complete cds
SCORE:3733.0
N.ALIGN:1
E-VAL:0.0
TITLE:gi|164692402|dbj|AK314794.1| Homo sapiens cDNA, FLJ95666, highly similar to Homo sapiens albumin (ALB), mRNA
SCORE:3676.0
N.ALIGN:1
E-VAL:0.0
TITLE:gi|52001696|gb|AY728024.1| Homo sapiens serum albumin precursor, mRNA, complete cds
SCORE:3630.0
N.ALIGN:1
E-VAL:0.0
TITLE:gi|123981021|gb|DQ891414.2| Synthetic construct clone IMAGE:100004044; FLH176691.01X; RZPDo839G03122D albumin (ALB) gene, encodes complete protein
SCORE:3626.0
N.ALIGN:1
E-VAL:0.0
TITLE:gi|123995824|gb|DQ894588.2| Synthetic construct Homo sapiens clone IMAGE:100009048; FLH176687.01L; RZPDo839G03121D albumin (ALB) gene, encodes complete protein
SCORE:3624.0
N.ALIGN:1
E-VAL:0.0
TITLE:gi|6013426|gb|AF190168.1|AF190168 Homo sapiens serum albumin precursor, mRNA, complete cds
SCORE:3615.0
N.ALIGN:1
E-VAL:0.0
TITLE:gi|332356379|gb|HQ537426.1| Homo sapiens albumin mRNA, partial cds
SCORE:3471.0
N.ALIGN:1
E-VAL:0.0
TITLE:gi|194391111|dbj|AK298461.1| Homo sapiens cDNA FLJ50830 complete cds, highly similar to Serum albumin precursor
SCORE:3084.0
N.ALIGN:2
E-VAL:0.0
TITLE:gi|763428|gb|U22961.1|HSU22961 Human mRNA clone with similarity to L-glycerol-3-phosphate:NAD oxidoreductase and albumin gene sequences
SCORE:2748.0
N.ALIGN:1
E-VAL:0.0
TITLE:gi|27692692|gb|BC041789.1| Homo sapiens albumin, mRNA (cDNA clone MGC:32888 IMAGE:4766983), complete cds
SCORE:2620.0
N.ALIGN:2
E-VAL:0.0
TITLE:gi|164694201|dbj|AK308044.1| Homo sapiens cDNA, FLJ97992
SCORE:2536.0
N.ALIGN:2
E-VAL:0.0
TITLE:gi|7770216|gb|AF119890.1|AF119890 Homo sapiens PRO2675 mRNA, complete cds
SCORE:2488.0
N.ALIGN:1
E-VAL:0.0
TITLE:gi|47118014|gb|AY544124.1| Homo sapiens growth-inhibiting protein 20 mRNA, complete cds
SCORE:2453.0
N.ALIGN:2
E-VAL:0.0
TITLE:gi|7959790|gb|AF116645.1|AF116645 Homo sapiens PRO1708 mRNA, complete cds
SCORE:2003.0
N.ALIGN:1
E-VAL:0.0
TITLE:gi|6650825|gb|AF118090.1|AF118090 Homo sapiens PRO2044 mRNA, complete cds
SCORE:1983.0
N.ALIGN:1
E-VAL:0.0
TITLE:gi|168745965|emb|CU691683.1| Synthetic construct Homo sapiens gateway clone IMAGE:100022016 3' read mRNA
SCORE:1951.0
N.ALIGN:1
E-VAL:0.0
TITLE:gi|168745964|emb|CU691682.1| Synthetic construct Homo sapiens gateway clone IMAGE:100022016 5' read mRNA
SCORE:1896.0
N.ALIGN:1
E-VAL:0.0
TITLE:gi|23241674|gb|BC035969.1| Homo sapiens albumin, mRNA (cDNA clone MGC:32581 IMAGE:4714468), complete cds
SCORE:1767.0
N.ALIGN:2
E-VAL:0.0
TITLE:gi|15679995|gb|BC014308.1|BC014308 Homo sapiens, clone IMAGE:3934797, mRNA, partial cds
SCORE:1571.0
N.ALIGN:1
E-VAL:0.0
TITLE:gi|649152310|gb|KJ905683.1| Synthetic construct Homo sapiens clone ccsbBroadEn_15353 ALB gene, encodes complete protein
SCORE:1387.0
N.ALIGN:2
E-VAL:0.0
TITLE:gi|168740012|emb|CU690239.1| Synthetic construct Homo sapiens gateway clone IMAGE:100022252 3' read ALB mRNA
SCORE:1182.0
N.ALIGN:1
E-VAL:0.0
TITLE:gi|37181745|gb|AY358313.1| Homo sapiens DNA66677 ALB (UNQ696) mRNA, partial cds
SCORE:1018.0
N.ALIGN:1
E-VAL:0.0
TITLE:gi|168740011|emb|CU690238.1| Synthetic construct Homo sapiens gateway clone IMAGE:100022252 5' read ALB mRNA
SCORE:982.0
N.ALIGN:2
E-VAL:0.0
TITLE:gi|6642751|gb|AF113014.1|AF113014 Homo sapiens clone FLB3512 mRNA sequence
SCORE:618.0
N.ALIGN:1
E-VAL:1.15187e-155
TITLE:gi|219842221|ref|NG_009291.1| Homo sapiens albumin (ALB), RefSeqGene on chromosome 4
SCORE:581.0
N.ALIGN:15
E-VAL:2.37628e-145
TITLE:gi|152112963|gb|EF649953.1| Homo sapiens albumin (ALB) gene, complete cds
SCORE:581.0
N.ALIGN:15
E-VAL:2.37628e-145
TITLE:gi|19387810|gb|AC108157.3| Homo sapiens BAC clone RP11-580P21 from 4, complete sequence
SCORE:581.0
N.ALIGN:16
E-VAL:2.37628e-145
TITLE:gi|178343|gb|M12523.1|HUMALBGC Human serum albumin (ALB) gene, complete cds
SCORE:581.0
N.ALIGN:15
E-VAL:2.37628e-145
TITLE:gi|1036032687|gb|AH007061.2| Homo sapiens chromosome 4 serum albumin (ALB) gene, partial cds; and serum albumin-alphafetoprotein intergenic spacer, partial sequence
SCORE:579.0
N.ALIGN:1
E-VAL:8.29405e-145
TITLE:gi|408407822|gb|AF116616.2| Homo sapiens PRO0998 mRNA, complete cds
SCORE:577.0
N.ALIGN:1
E-VAL:2.89491e-144
TITLE:gi|338858118|gb|AH002596.2|SEG_HUMALB Homo sapiens albumin (ALB) gene, partial cds
SCORE:573.0
N.ALIGN:2
E-VAL:3.52672e-143
TITLE:gi|1122817323|gb|DQ145726.2| Homo sapiens ARNT-interacting protein 2 (AINP2) mRNA, complete cds
SCORE:404.0
N.ALIGN:1
E-VAL:1.40699e-97
TITLE:gi|32428|emb|X51365.1| Homo sapiens mRNA for albumin (clone pHA19)
SCORE:232.0
N.ALIGN:1
E-VAL:6.8383e-51
TITLE:gi|1338686512|ref|NG_056261.1| Homo sapiens albumin (ALB) 5' regulatory region (LOC111832671) on chromosome 4
SCORE:221.0
N.ALIGN:1
E-VAL:1.23639e-47
TITLE:gi|7959920|gb|AF116711.1|AF116711 Homo sapiens PRO2646 mRNA, complete cds
SCORE:216.0
N.ALIGN:1
E-VAL:1.50624e-46
Align Title:gi|23307792|gb|AF542069.1| Homo sapiens serum albumin (HSA) mRNA, complete cds
Align Len: 2176
Score: 4352.0 Bits: 3925.42 E-val: 0.0
N.aligns:None Ident:2176 Pos.:2176 Gaps:0 Align len:2176
Strand: (None, None) Frame: (1, 1)
Query: TCTCTTCTGTCAACCCCACGCGCCTTTGGCACAATGAAGTGGGTAACCTTTATTTCCCTTCTTTTTCTCTTTAGCTCGGCTTATTCCAGGGGTGTGTTTCGTCGAGATGCACACAAGAGTGAGGTTGCTCATCGGTTTAAAGATTTGGGAGAAGAAAATTTCAAAGCCTTAGTGTTGATTGCCTTTGCTCAGTATCTTCAGCAGTGTCCATTTGAAGATCATGTAAAATTAGTGAATGAAGTAACTGAATTTGCAAAAACATGTGTTGCTGATGAGTCAGCTGAAAATTGTGACAAATCACTTCATACCCTTTTTGGAGACAAATTATGCACAGTTGCAACTCTTCGTGAAACCTATGGTGAAATGGCTGACTGCTGTGCAAAACAAGAACCTGAGAGAAATGAATGCTTCTTGCAACACAAAGATGACAACCCAAACCTCCCCCGATTGGTGAGACCAGAGGTTGATGTGATGTGCACTGCTTTTCATGACAATGAAGAGACATTTTTGAAAAAATACTTATATGAAATTGCCAGAAGACATCCTTACTTCTATGCCCCGGAACTCCTTTTCTTTGCTAAAAGGTATAAAGCTGCTTTTACAGAATGTTGCCAAGCTGCTGATAAGGCTGCCTGCCTGTTGCCAAAGCTCGATGAACTTCGGGATGAAGGGAAGGCTTCGTCTGCCAAACAGAGGCTCAAGTGTGCCAGTCTCCAAAAATTTGGAGAAAGAGCTTTCAAAGCATGGGCAGTAGCTCGCCTGAGCCAGAGATTTCCCAAAGCTGAGTTTGCAGAAGTTTCCAAGTTAGTGACAGATCTTACCAAAGTCCACACGGAATGCTGCCATGGAGATCTGCTTGAATGTGCTGATGACAGGGCGGACCTTGCCAAGTATATCTGTGAAAATCAAGATTCGATCTCCAGTAAACTGAAGGAATGCTGTGAAAAACCTCTGTTGGAAAAATCCCACTGCATTGCCGAAGTGGAAAATGATGAGATGCCTGCTGACTTGCCTTCATTAGCTGCTGATTTTGTTGAAAGTAAGGATGTTTGCAAAAACTATGCTGAGGCAAAGGATGTCTTCCTGGGCATGTTTTTGTATGAATATGCAAGAAGGCATCCTGATTACTCTGTCGTGCTGCTGCTGAGACTTGCCAAGACATATAAAACCACTCTAGAGAAGTGCTGTGCCGCTGCAGATCCTCATGAATGCTATGCCAAAGTGTTCGATGAATTTAAACCTCTTGTGGAAGAGCCTCAGAATTTAATCAAACAAAATTGTGAGCTTTTTGAGCAGCTTGGAGAGTACAAATTCCAGAATGCGCTATTAGTTCGTTACACCAAGAAAGTACCCCAAGTGTCAACTCCAACTCTTGTAGAGGTCTCAAGAAACCTAGGAAAAGTGGGCAGCAAATGTTGTAAACATCCTGAAGCAAAAAGAATGCCCTGTGCAGAAGACTATCTATCCGTGGTCCTGAACCAGTTATGTGTGTTGCATGAGAAAACGCCAGTAAGTGACAGAGTCACCAAATGCTGCACAGAATCCTTGGTGAACAGGCGACCATGCTTTTCAGCTCTGGAAGTCGATGAAACATACGTTCCCAAAGAGTTTAATGCTGAAACATTCACCTTCCATGCAGATATATGCACACTTTCTGAGAAGGAGAGACAAATCAAGAAACAAACTGCACTTGTTGAGCTTGTGAAACACAAGCCCAAGGCAACAAAAGAGCAACTGAAAGCTGTTATGGATGATTTCGCAGCTTTTGTAGAGAAGTGCTGCAAGGCTGACGATAAGGAGACCTGCTTTGCCGAGGAGGGTAAAAAACTTGTTGCTGCAAGTCGAGCTGCCTTAGGCTTATAACATCACATTTAAAAGCATCTCAGCCTACCATGAGAATAAGAGAAAGAAAATGAAGATCAAAAGCTTATTCATCTGTTTTTCTTTTTCGTTGGTGTAAAGCCAACACCCTGTCTAAAAAACATAAATTTCTTTAATCATTTTGCCTCTTTTCTCTGTGCTTCAATTAATAAAAAATGGAAAGAATCTAATAGAGTGGTACAGCACTGTTATTTTTCAAAGATGTGTTGCTATCCTGAAAATTCTGTAGGTTCTGTGGAAGTTCCAGTGTTCTCTCTTATTCCACTTCGGTAGAGGATTTCTAGTTTCTGTGGGC start: 1 end: 2176
Match: ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Subjc: TCTCTTCTGTCAACCCCACGCGCCTTTGGCACAATGAAGTGGGTAACCTTTATTTCCCTTCTTTTTCTCTTTAGCTCGGCTTATTCCAGGGGTGTGTTTCGTCGAGATGCACACAAGAGTGAGGTTGCTCATCGGTTTAAAGATTTGGGAGAAGAAAATTTCAAAGCCTTAGTGTTGATTGCCTTTGCTCAGTATCTTCAGCAGTGTCCATTTGAAGATCATGTAAAATTAGTGAATGAAGTAACTGAATTTGCAAAAACATGTGTTGCTGATGAGTCAGCTGAAAATTGTGACAAATCACTTCATACCCTTTTTGGAGACAAATTATGCACAGTTGCAACTCTTCGTGAAACCTATGGTGAAATGGCTGACTGCTGTGCAAAACAAGAACCTGAGAGAAATGAATGCTTCTTGCAACACAAAGATGACAACCCAAACCTCCCCCGATTGGTGAGACCAGAGGTTGATGTGATGTGCACTGCTTTTCATGACAATGAAGAGACATTTTTGAAAAAATACTTATATGAAATTGCCAGAAGACATCCTTACTTCTATGCCCCGGAACTCCTTTTCTTTGCTAAAAGGTATAAAGCTGCTTTTACAGAATGTTGCCAAGCTGCTGATAAGGCTGCCTGCCTGTTGCCAAAGCTCGATGAACTTCGGGATGAAGGGAAGGCTTCGTCTGCCAAACAGAGGCTCAAGTGTGCCAGTCTCCAAAAATTTGGAGAAAGAGCTTTCAAAGCATGGGCAGTAGCTCGCCTGAGCCAGAGATTTCCCAAAGCTGAGTTTGCAGAAGTTTCCAAGTTAGTGACAGATCTTACCAAAGTCCACACGGAATGCTGCCATGGAGATCTGCTTGAATGTGCTGATGACAGGGCGGACCTTGCCAAGTATATCTGTGAAAATCAAGATTCGATCTCCAGTAAACTGAAGGAATGCTGTGAAAAACCTCTGTTGGAAAAATCCCACTGCATTGCCGAAGTGGAAAATGATGAGATGCCTGCTGACTTGCCTTCATTAGCTGCTGATTTTGTTGAAAGTAAGGATGTTTGCAAAAACTATGCTGAGGCAAAGGATGTCTTCCTGGGCATGTTTTTGTATGAATATGCAAGAAGGCATCCTGATTACTCTGTCGTGCTGCTGCTGAGACTTGCCAAGACATATAAAACCACTCTAGAGAAGTGCTGTGCCGCTGCAGATCCTCATGAATGCTATGCCAAAGTGTTCGATGAATTTAAACCTCTTGTGGAAGAGCCTCAGAATTTAATCAAACAAAATTGTGAGCTTTTTGAGCAGCTTGGAGAGTACAAATTCCAGAATGCGCTATTAGTTCGTTACACCAAGAAAGTACCCCAAGTGTCAACTCCAACTCTTGTAGAGGTCTCAAGAAACCTAGGAAAAGTGGGCAGCAAATGTTGTAAACATCCTGAAGCAAAAAGAATGCCCTGTGCAGAAGACTATCTATCCGTGGTCCTGAACCAGTTATGTGTGTTGCATGAGAAAACGCCAGTAAGTGACAGAGTCACCAAATGCTGCACAGAATCCTTGGTGAACAGGCGACCATGCTTTTCAGCTCTGGAAGTCGATGAAACATACGTTCCCAAAGAGTTTAATGCTGAAACATTCACCTTCCATGCAGATATATGCACACTTTCTGAGAAGGAGAGACAAATCAAGAAACAAACTGCACTTGTTGAGCTTGTGAAACACAAGCCCAAGGCAACAAAAGAGCAACTGAAAGCTGTTATGGATGATTTCGCAGCTTTTGTAGAGAAGTGCTGCAAGGCTGACGATAAGGAGACCTGCTTTGCCGAGGAGGGTAAAAAACTTGTTGCTGCAAGTCGAGCTGCCTTAGGCTTATAACATCACATTTAAAAGCATCTCAGCCTACCATGAGAATAAGAGAAAGAAAATGAAGATCAAAAGCTTATTCATCTGTTTTTCTTTTTCGTTGGTGTAAAGCCAACACCCTGTCTAAAAAACATAAATTTCTTTAATCATTTTGCCTCTTTTCTCTGTGCTTCAATTAATAAAAAATGGAAAGAATCTAATAGAGTGGTACAGCACTGTTATTTTTCAAAGATGTGTTGCTATCCTGAAAATTCTGTAGGTTCTGTGGAAGTTCCAGTGTTCTCTCTTATTCCACTTCGGTAGAGGATTTCTAGTTTCTGTGGGC start: 1 end: 2176
Align Title:gi|1519245814|ref|NM_000477.7| Homo sapiens albumin (ALB), mRNA
Align Len: 2285
Score: 4305.0 Bits: 3883.04 E-val: 0.0
N.aligns:None Ident:2168 Pos.:2168 Gaps:1 Align len:2177
Strand: (None, None) Frame: (1, 1)
Query: TCTCTTCTGTCAACCCCACGCGCCTTTGGCACAATGAAGTGGGTAACCTTTATTTCCCTTCTTTTTCTCTTTAGCTCGGCTTATTCCAGGGGTGTGTTTCGTCGAGATGCACACAAGAGTGAGGTTGCTCATCGGTTTAAAGATTTGGGAGAAGAAAATTTCAAAGCCTTAGTGTTGATTGCCTTTGCTCAGTATCTTCAGCAGTGTCCATTTGAAGATCATGTAAAATTAGTGAATGAAGTAACTGAATTTGCAAAAACATGTGTTGCTGATGAGTCAGCTGAAAATTGTGACAAATCACTTCATACCCTTTTTGGAGACAAATTATGCACAGTTGCAACTCTTCGTGAAACCTATGGTGAAATGGCTGACTGCTGTGCAAAACAAGAACCTGAGAGAAATGAATGCTTCTTGCAACACAAAGATGACAACCCAAACCTCCCCCGATTGGTGAGACCAGAGGTTGATGTGATGTGCACTGCTTTTCATGACAATGAAGAGACATTTTTGAAAAAATACTTATATGAAATTGCCAGAAGACATCCTTACTTCTATGCCCCGGAACTCCTTTTCTTTGCTAAAAGGTATAAAGCTGCTTTTACAGAATGTTGCCAAGCTGCTGATAAGGCTGCCTGCCTGTTGCCAAAGCTCGATGAACTTCGGGATGAAGGGAAGGCTTCGTCTGCCAAACAGAGGCTCAAGTGTGCCAGTCTCCAAAAATTTGGAGAAAGAGCTTTCAAAGCATGGGCAGTAGCTCGCCTGAGCCAGAGATTTCCCAAAGCTGAGTTTGCAGAAGTTTCCAAGTTAGTGACAGATCTTACCAAAGTCCACACGGAATGCTGCCATGGAGATCTGCTTGAATGTGCTGATGACAGGGCGGACCTTGCCAAGTATATCTGTGAAAATCAAGATTCGATCTCCAGTAAACTGAAGGAATGCTGTGAAAAACCTCTGTTGGAAAAATCCCACTGCATTGCCGAAGTGGAAAATGATGAGATGCCTGCTGACTTGCCTTCATTAGCTGCTGATTTTGTTGAAAGTAAGGATGTTTGCAAAAACTATGCTGAGGCAAAGGATGTCTTCCTGGGCATGTTTTTGTATGAATATGCAAGAAGGCATCCTGATTACTCTGTCGTGCTGCTGCTGAGACTTGCCAAGACATATAAAACCACTCTAGAGAAGTGCTGTGCCGCTGCAGATCCTCATGAATGCTATGCCAAAGTGTTCGATGAATTTAAACCTCTTGTGGAAGAGCCTCAGAATTTAATCAAACAAAATTGTGAGCTTTTTGAGCAGCTTGGAGAGTACAAATTCCAGAATGCGCTATTAGTTCGTTACACCAAGAAAGTACCCCAAGTGTCAACTCCAACTCTTGTAGAGGTCTCAAGAAACCTAGGAAAAGTGGGCAGCAAATGTTGTAAACATCCTGAAGCAAAAAGAATGCCCTGTGCAGAAGACTATCTATCCGTGGTCCTGAACCAGTTATGTGTGTTGCATGAGAAAACGCCAGTAAGTGACAGAGTCACCAAATGCTGCACAGAATCCTTGGTGAACAGGCGACCATGCTTTTCAGCTCTGGAAGTCGATGAAACATACGTTCCCAAAGAGTTTAATGCTGAAACATTCACCTTCCATGCAGATATATGCACACTTTCTGAGAAGGAGAGACAAATCAAGAAACAAACTGCACTTGTTGAGCTTGTGAAACACAAGCCCAAGGCAACAAAAGAGCAACTGAAAGCTGTTATGGATGATTTCGCAGCTTTTGTAGAGAAGTGCTGCAAGGCTGACGATAAGGAGACCTGCTTTGCCGAGGAGGGTAAAAAACTTGTTGCTGCAAGTCGAGCTGCCTTAGGCTTATAACATCACATTTAAAAGCATCTCAGCCTACCATGAGAATAAGAGAAAGAAAATGAAGATCAAAAGCTTATTCATCTGTTTTTCTTTTTCGTTGGTGTAAAGCCAACACCCTGTCTAAAAAACATAAATTTCTTTAATCATTTTGCCTCTTTTCTCTGTGCTTCAATTAATAAAAAATGGAAAGAATCTAATAGAGTGGTACAGCACTGTTATTTTTCAAAGATGTGTTGCTATCCTGAAAATTCTGTAGGTTCTGTGGAAGTTCCAGTGTTCTCTCTTATTCCACTTCGGTAGAGGATTTCTAGTTTC-TGTGGGC start: 1 end: 2176
Match: ||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||
Subjc: TCTCTTCTGTCAACCCCACACGCCTTTGGCACAATGAAGTGGGTAACCTTTATTTCCCTTCTTTTTCTCTTTAGCTCGGCTTATTCCAGGGGTGTGTTTCGTCGAGATGCACACAAGAGTGAGGTTGCTCATCGGTTTAAAGATTTGGGAGAAGAAAATTTCAAAGCCTTGGTGTTGATTGCCTTTGCTCAGTATCTTCAGCAGTGTCCATTTGAAGATCATGTAAAATTAGTGAATGAAGTAACTGAATTTGCAAAAACATGTGTTGCTGATGAGTCAGCTGAAAATTGTGACAAATCACTTCATACCCTTTTTGGAGACAAATTATGCACAGTTGCAACTCTTCGTGAAACCTATGGTGAAATGGCTGACTGCTGTGCAAAACAAGAACCTGAGAGAAATGAATGCTTCTTGCAACACAAAGATGACAACCCAAACCTCCCCCGATTGGTGAGACCAGAGGTTGATGTGATGTGCACTGCTTTTCATGACAATGAAGAGACATTTTTGAAAAAATACTTATATGAAATTGCCAGAAGACATCCTTACTTTTATGCCCCGGAACTCCTTTTCTTTGCTAAAAGGTATAAAGCTGCTTTTACAGAATGTTGCCAAGCTGCTGATAAAGCTGCCTGCCTGTTGCCAAAGCTCGATGAACTTCGGGATGAAGGGAAGGCTTCGTCTGCCAAACAGAGACTCAAGTGTGCCAGTCTCCAAAAATTTGGAGAAAGAGCTTTCAAAGCATGGGCAGTAGCTCGCCTGAGCCAGAGATTTCCCAAAGCTGAGTTTGCAGAAGTTTCCAAGTTAGTGACAGATCTTACCAAAGTCCACACGGAATGCTGCCATGGAGATCTGCTTGAATGTGCTGATGACAGGGCGGACCTTGCCAAGTATATCTGTGAAAATCAAGATTCGATCTCCAGTAAACTGAAGGAATGCTGTGAAAAACCTCTGTTGGAAAAATCCCACTGCATTGCCGAAGTGGAAAATGATGAGATGCCTGCTGACTTGCCTTCATTAGCTGCTGATTTTGTTGAAAGTAAGGATGTTTGCAAAAACTATGCTGAGGCAAAGGATGTCTTCCTGGGCATGTTTTTGTATGAATATGCAAGAAGGCATCCTGATTACTCTGTCGTGCTGCTGCTGAGACTTGCCAAGACATATGAAACCACTCTAGAGAAGTGCTGTGCCGCTGCAGATCCTCATGAATGCTATGCCAAAGTGTTCGATGAATTTAAACCTCTTGTGGAAGAGCCTCAGAATTTAATCAAACAAAATTGTGAGCTTTTTGAGCAGCTTGGAGAGTACAAATTCCAGAATGCGCTATTAGTTCGTTACACCAAGAAAGTACCCCAAGTGTCAACTCCAACTCTTGTAGAGGTCTCAAGAAACCTAGGAAAAGTGGGCAGCAAATGTTGTAAACATCCTGAAGCAAAAAGAATGCCCTGTGCAGAAGACTATCTATCCGTGGTCCTGAACCAGTTATGTGTGTTGCATGAGAAAACGCCAGTAAGTGACAGAGTCACCAAATGCTGCACAGAATCCTTGGTGAACAGGCGACCATGCTTTTCAGCTCTGGAAGTCGATGAAACATACGTTCCCAAAGAGTTTAATGCTGAAACATTCACCTTCCATGCAGATATATGCACACTTTCTGAGAAGGAGAGACAAATCAAGAAACAAACTGCACTTGTTGAGCTCGTGAAACACAAGCCCAAGGCAACAAAAGAGCAACTGAAAGCTGTTATGGATGATTTCGCAGCTTTTGTAGAGAAGTGCTGCAAGGCTGACGATAAGGAGACCTGCTTTGCCGAGGAGGGTAAAAAACTTGTTGCTGCAAGTCAAGCTGCCTTAGGCTTATAACATCACATTTAAAAGCATCTCAGCCTACCATGAGAATAAGAGAAAGAAAATGAAGATCAAAAGCTTATTCATCTGTTTTTCTTTTTCGTTGGTGTAAAGCCAACACCCTGTCTAAAAAACATAAATTTCTTTAATCATTTTGCCTCTTTTCTCTGTGCTTCAATTAATAAAAAATGGAAAGAATCTAATAGAGTGGTACAGCACTGTTATTTTTCAAAGATGTGTTGCTATCCTGAAAATTCTGTAGGTTCTGTGGAAGTTCCAGTGTTCTCTCTTATTCCACTTCGGTAGAGGATTTCTAGTTTCTTGTGGGC start: 9 end: 2185
Align Title:gi|28591|emb|V00495.1| H.sapiens mRNA for serum albumin
Align Len: 2251
Score: 4253.0 Bits: 3836.15 E-val: 0.0
N.aligns:None Ident:2159 Pos.:2159 Gaps:4 Align len:2177
Strand: (None, None) Frame: (1, 1)
Query: TCTCTTCTGTCAACCCCACGCGCCTTTGGCACAATGAAGTGGGTAACCTTTATTTCCCTTCTTTTTCTCTTTAGCTCGGCTTATTCCAGGGGTGTGTTTCGTCGAGATGCACACAAGAGTGAGGTTGCTCATCGGTTTAAAGATTTGGGAGAAGAAAATTTCAAAGCCTTAGTGTTGATTGCCTTTGCTCAGTATCTTCAGCAGTGTCCATTTGAAGATCATGTAAAATTAGTGAATGAAGTAACTGAATTTGCAAAAACATGTGTTGCTGATGAGTCAGCTGAAAATTGTGACAAATCACTTCATACCCTTTTTGGAGACAAATTATGCACAGTTGCAACTCTTCGTGAAACCTATGGTGAAATGGCTGACTGCTGTGCAAAACAAGAACCTGAGAGAAATGAATGCTTCTTGCAACACAAAGATGACAACCCAAACCTCCCCCGATTGGTGAGACCAGAGGTTGATGTGATGTGCACTGCTTTTCATGACAATGAAGAGACATTTTTGAAAAAATACTTATATGAAATTGCCAGAAGACATCCTTACTTCTATGCCCCGGAACTCCTTTTCTTTGCTAAAAGGTATAAAGCTGCTTTTACAGAATGTTGCCAAGCTGCTGATAAGGCTGCCTGCCTGTTGCCAAAGCTCGATGAACTTCGGGATGAAGGGAAGGCTTCGTCTGCCAAACAGAGGCTCAAGTGTGCCAGTCTCCAAAAATTTGGAGAAAGAGCTTTCAAAGCATGGGCAGTAGCTCGCCTGAGCCAGAGATTTCCCAAAGCTGAGTTTGCAGAAGTTTCCAAGTTAGTGACAGATCTTACCAAAGTCCACACGGAATGCTGCCATGGAGATCTGCTTGAATGTGCTGATGACAGGGCGGACCTTGCCAAGTATATCTGTGAAAATCAAGATTCGATCTCCAGTAAACTGAAGGAATGCTGTGAAAAACCTCTGTTGGAAAAATCCCACTGCATTGCCGAAGTGGAAAATGATGAGATGCCTGCTGACTTGCCTTCATTAGCTGCTGATTTTGTTGAAAGTAAGGATGTTTGCAAAAACTATGCTGAGGCAAAGGATGTCTTCCTGGGCATGTTTTTGTATGAATATGCAAGAAGGCATCCTGATTACTCTGTCGTGCTGCTGCTGAGACTTGCCAAGACATATAAAACCACTCTAGAGAAGTGCTGTGCCGCTGCAGATCCTCATGAATGCTATGCCAAAGTGTTCGATGAATTTAAACCTCTTGTGGAAGAGCCTCAGAATTTAATCAAACAAAATTGTGAGCTTTTTGAGCAGCTTGGAGAGTACAAATTCCAGAATGCGCTATTAGTTCGTTACACCAAGAAAGTACCCCAAGTGTCAACTCCAACTCTTGTAGAGGTCTCAAGAAACCTAGGAAAAGTGGGCAGCAAATGTTGTAAACATCCTGAAGCAAAAAGAATGCCCTGTGCAGAAGACTATCTATCCGTGGTCCTGAACCAGTTATGTGTGTTGCATGAGAAAACGCCAGTAAGTGACAGAGTCACCAAATGCTGCACAGAATCCTTGGTGAACAGGCGACCATGCTTTTCAGCTCTGGAAGTCGATGAAACATACGTTCCCAAAGAGTTTAATGCTGAAACATTCACCTTCCATGCAGATATATGCACACTTTCTGAGAAGGAGAGACAAATCAAGAAACAAACTGCACTTGTTGAGCTTGTGAAACACAAGCCCAAGGCAACAAAAGAGCAACTGAAAGCTGTTATGGATGATTTCGCAGCTTTTGTAGAGAAGTGCTGCAAGGCTGACGATAAGGAGACCTGCTTTGCCGAGGAGGGTAAAAAACTTGTTGCTGCAAGTCGAGCTGCCTTAGGCTTATAACATC-ACATTTAAAAGCATCTCAGCCTACCATGAGAATAAGAGAAAGAAAATGAAGATCAAAAGCTTATTCATCTGTTTTTCTTTTTCGTTGGTGTAAAGCCAACACCCTGTCTAAAAAACATAAATTTCTTTAATCATTTTGCCTCTTTTCTCTGTGCTTCAATTAATAAAAAATGGAAAGAATCTAATAGAGTGGTACAGCACTGTTATTTTTCAAAGATGTGTTGCTATCCTGAAAATTCTGTAGGTTCTGTGGAAGTTCCAGTGTTCTCTCTTATTCCACTTCGGTAGAGGATTTCTAGTTTCTGTGGGC start: 1 end: 2176
Match: ||||||||||||||||||||| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| ||||| |||||||||||||||||||||||||||||||||||||||||||||||||| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| ||||||||||||||||||||||| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Subjc: TCTCTTCTGTCAACCCCACGC--CTTTGGCACAATGAAGTGGGTAACCTTTATTTCCCTTCTTTTTCTCTTTAGCTCGGCTTATTCCAGGGGTGTGTTTCGTCGAGATGCACACAAGAGTGAGGTTGCTCATCGGTTTAAAGATTTGGGAGAAGAAAATTTCAAAGCCTTGGTGTTGATTGCCTTTGCTCAGTATCTTCAGCAGTGTCCATTTGAAGATCATGTAAAATTAGTGAATGAAGTAACTGAATTTGCAAAAACATGTGTAGCTGATGAGTCAGCTGAAAATTGTGACAAATCACTTCATACCCTTTTTGGAGACAAATTATGCACAGTTGCAACTCTTCGTGAAACCTATGGTGAAATGGCTGACTGCTGTGCAAAACAAGAACCTGAGAGAAATGAATGCTTCTTGCAACACAAAGATGACAACCCAAACCTCCCCCGATTGGTGAGACCAGAGGTTGATGTGATGTGCACTGCTTTTCATGACAATGAAGAGACATTTTTGAAAAAATACTTATATGAAATTGCCAGAAGACATCCTTACTTTTATGCCCCGGAACTCCTTTTCTTTGCTAAAAGGTATAAAGCTGCTTTTACAGAATGTTGCCAAGCTGCTGATAAAGCTGCCTGCCTGTTGCCAAAGCTCGATGAACTTCGGGATGAAGGGAAGGCTTCGTCTGCCAAACAGAGACTCAAATGTGCCAGTCTCCAAAAATTTGGAGAAAGAGCTTTCAAAGCATGGGCAGTGGCTCGCCTGAGCCAGAGATTTCCCAAAGCTGAGTTTGCAGAAGTTTCCAAGTTAGTGACAGATCTTACCAAAGTCCACACGGAATGCTGCCATGGAGATCTGCTTGAATGTGCTGATGACAGGGCGGACCTTGCCAAGTATATCTGTGAAAATCAGGATTCGATCTCCAGTAAACTGAAGGAATGCTGTGAAAAACCTCTGTTGGAAAAATCCCACTGCATTGCCGAAGTGGAAAATGATGAGATGCCTGCTGACTTGCCTTCATTAGCTGCTGATTTTGTTGAAAGTAAGGATGTTTGCAAAAACTATGCTGAGGCAAAGGATGTCTTCCTGGGCATGTTTTTGTATGAATATGCAAGAAGGCATCCTGATTACTCTGTCGTGCTGCTGCTGAGACTTGCCAAGACATATGAAACCACTCTAGAGAAGTGCTGTGCCGCTGCAGATCCTCATGAATGCTATGCCAAAGTGTTCGATGAATTTAAACCTCTTGTGGAAGAGCCTCAGAATTTAATCAAACAAAACTGTGAGCTTTTTAAGCAGCTTGGAGAGTACAAATTCCAGAATGCGCTATTAGTTCGTTACACCAAGAAAGTACCCCAAGTGTCAACTCCAACTCTTGTAGAGGTCTCAAGAAACCTAGGAAAAGTGGGCAGCAAATGTTGTAAACATCCTGAAGCAAAAAGAATGCCCTGTGCAGAAGACTATCTATCCGTGGTCCTGAACCAGTTATGTGTGTTGCATGAGAAAACGCCAGTAAGTGACAGAGTCACAAAATGCTGCACAGAGTCCTTGGTGAACAGGCGACCATGCTTTTCAGCTCTGGAAGTCGATGAAACATACGTTCCCAAAGAGTTTAATGCTGAAACATTCACCTTCCATGCAGATATATGCACACTTTCTGAGAAGGAGAGACAAATCAAGAAACAAACTGCACTTGTTGAGCTTGTGAAACACAAGCCCAAGGCAACAAAAGAGCAACTGAAAGCTGTTATGGATGATTTCGCAGCTTTTGTAGAGAAGTGCTGCAAGGCTGACGATAAGGAGACCTGCTTTGCCGAGGAGGGTAAAAAACTTGTTGCTGCAAGTCAAGCTGCCTTAGGCTTATAACATCTACATTTAAAAGCATCTCAGCCTACCATGAGAATAAGAGAAAGAAAATGAAGATCAAAAGCTTATTCATCTG-TTTTCTTTTTCGTTGGTGTAAAGCCAACACCCTGTCTAAAAAACATAAATTTCTTTAATCATTTTGCCTCTTTTCTCTGTGCTTCAATTAATAAAAAATGGAAAGAATCTAATAGAGTGGTACAGCACTGTTATTTTTCAAAGATGTGTTGCTATCCTGAAAATTCTGTAGGTTCTGTGGAAGTTCCAGTGTTCTCTCTTATTCCACTTCGGTAGAGGATTTCTAGTTTCTGTGGGC start: 45 end: 2218
Score: 103.0 Bits: 94.1598 E-val: 1.32244e-15
N.aligns:None Ident:56 Pos.:56 Gaps:2 Align len:58
Strand: (None, None) Frame: (1, -1)
Query: TGACAATGAAGAGACATTTTTGAAAAAATACTTATATGAAATTGCCAGAAGACATCCT start: 489 end: 546
Match: ||||| |||||||||||||||||||||||||||||||||||||||||||||||||||
Subjc: TGACA--GAAGAGACATTTTTGAAAAAATACTTATATGAAATTGCCAGAAGACATCCT start: 56 end: 1
Align Title:gi|7770116|gb|AF119840.1|AF119840 Homo sapiens PRO0903 mRNA, complete cds
Align Len: 2720
Score: 4062.0 Bits: 3663.93 E-val: 0.0
N.aligns:None Ident:2043 Pos.:2043 Gaps:0 Align len:2051
Strand: (None, None) Frame: (1, 1)
Query: TCTCTTCTGTCAACCCCACGCGCCTTTGGCACAATGAAGTGGGTAACCTTTATTTCCCTTCTTTTTCTCTTTAGCTCGGCTTATTCCAGGGGTGTGTTTCGTCGAGATGCACACAAGAGTGAGGTTGCTCATCGGTTTAAAGATTTGGGAGAAGAAAATTTCAAAGCCTTAGTGTTGATTGCCTTTGCTCAGTATCTTCAGCAGTGTCCATTTGAAGATCATGTAAAATTAGTGAATGAAGTAACTGAATTTGCAAAAACATGTGTTGCTGATGAGTCAGCTGAAAATTGTGACAAATCACTTCATACCCTTTTTGGAGACAAATTATGCACAGTTGCAACTCTTCGTGAAACCTATGGTGAAATGGCTGACTGCTGTGCAAAACAAGAACCTGAGAGAAATGAATGCTTCTTGCAACACAAAGATGACAACCCAAACCTCCCCCGATTGGTGAGACCAGAGGTTGATGTGATGTGCACTGCTTTTCATGACAATGAAGAGACATTTTTGAAAAAATACTTATATGAAATTGCCAGAAGACATCCTTACTTCTATGCCCCGGAACTCCTTTTCTTTGCTAAAAGGTATAAAGCTGCTTTTACAGAATGTTGCCAAGCTGCTGATAAGGCTGCCTGCCTGTTGCCAAAGCTCGATGAACTTCGGGATGAAGGGAAGGCTTCGTCTGCCAAACAGAGGCTCAAGTGTGCCAGTCTCCAAAAATTTGGAGAAAGAGCTTTCAAAGCATGGGCAGTAGCTCGCCTGAGCCAGAGATTTCCCAAAGCTGAGTTTGCAGAAGTTTCCAAGTTAGTGACAGATCTTACCAAAGTCCACACGGAATGCTGCCATGGAGATCTGCTTGAATGTGCTGATGACAGGGCGGACCTTGCCAAGTATATCTGTGAAAATCAAGATTCGATCTCCAGTAAACTGAAGGAATGCTGTGAAAAACCTCTGTTGGAAAAATCCCACTGCATTGCCGAAGTGGAAAATGATGAGATGCCTGCTGACTTGCCTTCATTAGCTGCTGATTTTGTTGAAAGTAAGGATGTTTGCAAAAACTATGCTGAGGCAAAGGATGTCTTCCTGGGCATGTTTTTGTATGAATATGCAAGAAGGCATCCTGATTACTCTGTCGTGCTGCTGCTGAGACTTGCCAAGACATATAAAACCACTCTAGAGAAGTGCTGTGCCGCTGCAGATCCTCATGAATGCTATGCCAAAGTGTTCGATGAATTTAAACCTCTTGTGGAAGAGCCTCAGAATTTAATCAAACAAAATTGTGAGCTTTTTGAGCAGCTTGGAGAGTACAAATTCCAGAATGCGCTATTAGTTCGTTACACCAAGAAAGTACCCCAAGTGTCAACTCCAACTCTTGTAGAGGTCTCAAGAAACCTAGGAAAAGTGGGCAGCAAATGTTGTAAACATCCTGAAGCAAAAAGAATGCCCTGTGCAGAAGACTATCTATCCGTGGTCCTGAACCAGTTATGTGTGTTGCATGAGAAAACGCCAGTAAGTGACAGAGTCACCAAATGCTGCACAGAATCCTTGGTGAACAGGCGACCATGCTTTTCAGCTCTGGAAGTCGATGAAACATACGTTCCCAAAGAGTTTAATGCTGAAACATTCACCTTCCATGCAGATATATGCACACTTTCTGAGAAGGAGAGACAAATCAAGAAACAAACTGCACTTGTTGAGCTTGTGAAACACAAGCCCAAGGCAACAAAAGAGCAACTGAAAGCTGTTATGGATGATTTCGCAGCTTTTGTAGAGAAGTGCTGCAAGGCTGACGATAAGGAGACCTGCTTTGCCGAGGAGGGTAAAAAACTTGTTGCTGCAAGTCGAGCTGCCTTAGGCTTATAACATCACATTTAAAAGCATCTCAGCCTACCATGAGAATAAGAGAAAGAAAATGAAGATCAAAAGCTTATTCATCTGTTTTTCTTTTTCGTTGGTGTAAAGCCAACACCCTGTCTAAAAAACATAAATTTCTTTAATCATTTTGCCTCTTTTCTCTGTGCTTCAATTAATAAAAAATGGAAAGAATCTAA start: 1 end: 2051
Match: ||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Subjc: TCTCTTCTGTCAACCCCACACGCCTTTGGCACAATGAAGTGGGTAACCTTTATTTCCCTTCTTTTTCTCTTTAGCTCGGCTTATTCCAGGGGTGTGTTTCGTCGAGATGCACACAAGAGTGAGGTTGCTCATCGGTTTAAAGATTTGGGAGAAGAAAATTTCAAAGCCTTGGTGTTGATTGCCTTTGCTCAGTATCTTCAGCAGTGTCCATTTGAAGATCATGTAAAATTAGTGAATGAAGTAACTGAATTTGCAAAAACATGTGTTGCTGATGAGTCAGCTGAAAATTGTGACAAATCACTTCATACCCTTTTTGGAGACAAATTATGCACAGTTGCAACTCTTCGTGAAACCTATGGTGAAATGGCTGACTGCTGTGCAAAACAAGAACCTGAGAGAAATGAATGCTTCTTGCAACACAAAGATGACAACCCAAACCTCCCCCGATTGGTGAGACCAGAGGTTGATGTGATGTGCACTGCTTTTCATGACAATGAAGAGACATTTTTGAAAAAATACTTATATGAAATTGCCAGAAGACATCCTTACTTTTATGCCCCGGAACTCCTTTTCTTTGCTAAAAGGTATAAAGCTGCTTTTACAGAATGTTGCCAAGCTGCTGATAAAGCTGCCTGCCTGTTGCCAAAGCTCGATGAACTTCGGGATGAAGGGAAGGCTTCGTCTGCCAAACAGAGACTCAAGTGTGCCAGTCTCCAAAAATTTGGAGAAAGAGCTTTCAAAGCATGGGCAGTAGCTCGCCTGAGCCAGAGATTTCCCAAAGCTGAGTTTGCAGAAGTTTCCAAGTTAGTGACAGATCTTACCAAAGTCCACACGGAATGCTGCCATGGAGATCTGCTTGAATGTGCTGATGACAGGGCGGACCTTGCCAAGTATATCTGTGAAAATCAAGATTCGATCTCCAGTAAACTGAAGGAATGCTGTGAAAAACCTCTGTTGGAAAAATCCCACTGCATTGCCGAAGTGGAAAATGATGAGATGCCTGCTGACTTGCCTTCATTAGCTGCTGATTTTGTTGAAAGTAAGGATGTTTGCAAAAACTATGCTGAGGCAAAGGATGTCTTCCTGGGCATGTTTTTGTATGAATATGCAAGAAGGCATCCTGATTACTCTGTCGTGCTGCTGCTGAGACTTGCCAAGACATATGAAACCACTCTAGAGAAGTGCTGTGCCGCTGCAGATCCTCATGAATGCTATGCCAAAGTGTTCGATGAATTTAAACCTCTTGTGGAAGAGCCTCAGAATTTAATCAAACAAAATTGTGAGCTTTTTGAGCAGCTTGGAGAGTACAAATTCCAGAATGCGCTATTAGTTCGTTACACCAAGAAAGTACCCCAAGTGTCAACTCCAACTCTTGTAGAGGTCTCAAGAAACCTAGGAAAAGTGGGCAGCAAATGTTGTAAACATCCTGAAGCAAAAAGAATGCCCTGTGCAGAAGACTATCTATCCGTGGTCCTGAACCAGTTATGTGTGTTGCATGAGAAAACGCCAGTAAGTGACAGAGTCACCAAATGCTGCACAGAATCCTTGGTGAACAGGCGACCATGCTTTTCAGCTCTGGAAGTCGATGAAACATACGTTCCCAAAGAGTTTAATGCTGAAACATTCACCTTCCATGCAGATATATGCACACTTTCTGAGAAGGAGAGACAAATCAAGAAACAAACTGCACTTGTTGAGCTCGTGAAACACAAGCCCAAGGCAACAAAAGAGCAACTGAAAGCTGTTATGGATGATTTCGCAGCTTTTGTAGAGAAGTGCTGCAAGGCTGACGATAAGGAGACCTGCTTTGCCGAGGAGGGTAAAAAACTTGTTGCTGCAAGTCAAGCTGCCTTAGGCTTATAACATCACATTTAAAAGCATCTCAGCCTACCATGAGAATAAGAGAAAGAAAATGAAGATCAAAAGCTTATTCATCTGTTTTTCTTTTTCGTTGGTGTAAAGCCAACACCCTGTCTAAAAAACATAAATTTCTTTAATCATTTTGCCTCTTTTCTCTGTGCTTCAATTAATAAAAAATGGAAAGAATCTAA start: 649 end: 2699
Align Title:gi|25058738|gb|BC039235.1| Homo sapiens albumin, mRNA (cDNA clone IMAGE:4768004), containing frame-shift errors
Align Len: 2085
Score: 4056.0 Bits: 3658.52 E-val: 0.0
N.aligns:None Ident:2042 Pos.:2042 Gaps:1 Align len:2050
Strand: (None, None) Frame: (1, 1)
Query: TCTCTTCTGTCAACCCCACGCGCCTTTGGCACAATGAAGTGGGTAACCTTTATTTCCCTTCTTTTTCTCTTTAGCTCGGCTTATTCCAGGGGTGTGTTTCGTCGAGATGCACACAAGAGTGAGGTTGCTCATCGGTTTAAAGATTTGGGAGAAGAAAATTTCAAAGCCTTAGTGTTGATTGCCTTTGCTCAGTATCTTCAGCAGTGTCCATTTGAAGATCATGTAAAATTAGTGAATGAAGTAACTGAATTTGCAAAAACATGTGTTGCTGATGAGTCAGCTGAAAATTGTGACAAATCACTTCATACCCTTTTTGGAGACAAATTATGCACAGTTGCAACTCTTCGTGAAACCTATGGTGAAATGGCTGACTGCTGTGCAAAACAAGAACCTGAGAGAAATGAATGCTTCTTGCAACACAAAGATGACAACCCAAACCTCCCCCGATTGGTGAGACCAGAGGTTGATGTGATGTGCACTGCTTTTCATGACAATGAAGAGACATTTTTGAAAAAATACTTATATGAAATTGCCAGAAGACATCCTTACTTCTATGCCCCGGAACTCCTTTTCTTTGCTAAAAGGTATAAAGCTGCTTTTACAGAATGTTGCCAAGCTGCTGATAAGGCTGCCTGCCTGTTGCCAAAGCTCGATGAACTTCGGGATGAAGGGAAGGCTTCGTCTGCCAAACAGAGGCTCAAGTGTGCCAGTCTCCAAAAATTTGGAGAAAGAGCTTTCAAAGCATGGGCAGTAGCTCGCCTGAGCCAGAGATTTCCCAAAGCTGAGTTTGCAGAAGTTTCCAAGTTAGTGACAGATCTTACCAAAGTCCACACGGAATGCTGCCATGGAGATCTGCTTGAATGTGCTGATGACAGGGCGGACCTTGCCAAGTATATCTGTGAAAATCAAGATTCGATCTCCAGTAAACTGAAGGAATGCTGTGAAAAACCTCTGTTGGAAAAATCCCACTGCATTGCCGAAGTGGAAAATGATGAGATGCCTGCTGACTTGCCTTCATTAGCTGCTGATTTTGTTGAAAGTAAGGATGTTTGCAAAAACTATGCTGAGGCAAAGGATGTCTTCCTGGGCATGTTTTTGTATGAATATGCAAGAAGGCATCCTGATTACTCTGTCGTGCTGCTGCTGAGACTTGCCAAGACATATAAAACCACTCTAGAGAAGTGCTGTGCCGCTGCAGATCCTCATGAATGCTATGCCAAAGTGTTCGATGAATTTAAACCTCTTGTGGAAGAGCCTCAGAATTTAATCAAACAAAATTGTGAGCTTTTTGAGCAGCTTGGAGAGTACAAATTCCAGAATGCGCTATTAGTTCGTTACACCAAGAAAGTACCCCAAGTGTCAACTCCAACTCTTGTAGAGGTCTCAAGAAACCTAGGAAAAGTGGGCAGCAAATGTTGTAAACATCCTGAAGCAAAAAGAATGCCCTGTGCAGAAGACTATCTATCCGTGGTCCTGAACCAGTTATGTGTGTTGCATGAGAAAACGCCAGTAAGTGACAGAGTCACCAAATGCTGCACAGAATCCTTGGTGAACAGGCGACCATGCTTTTCAGCTCTGGAAGTCGATGAAACATACGTTCCCAAAGAGTTTAATGCTGAAACATTCACCTTCCATGCAGATATATGCACACTTTCTGAGAAGGAGAGACAAATCAAGAAACAAACTGCACTTGTTGAGCTTGTGAAACACAAGCCCAAGGCAACAAAAGAGCAACTGAAAGCTGTTATGGATGATTTCGCAGCTTTTGTAGAGAAGTGCTGCAAGGCTGACGATAAGGAGACCTGCTTTGCCGAGGAGGGT-AAAAAACTTGTTGCTGCAAGTCGAGCTGCCTTAGGCTTATAACATCACATTTAAAAGCATCTCAGCCTACCATGAGAATAAGAGAAAGAAAATGAAGATCAAAAGCTTATTCATCTGTTTTTCTTTTTCGTTGGTGTAAAGCCAACACCCTGTCTAAAAAACATAAATTTCTTTAATCATTTTGCCTCTTTTCTCTGTGCTTCAATTAATAAAAAATGGAAAGAATCT start: 1 end: 2049
Match: ||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Subjc: TCTCTTCTGTCAACCCCACACGCCTTTGGCACAATGAAGTGGGTAACCTTTATTTCCCTTCTTTTTCTCTTTAGCTCGGCTTATTCCAGGGGTGTGTTTCGTCGAGATGCACACAAGAGTGAGGTTGCTCATCGGTTTAAAGATTTGGGAGAAGAAAATTTCAAAGCCTTGGTGTTGATTGCCTTTGCTCAGTATCTTCAGCAGTGTCCATTTGAAGATCATGTAAAATTAGTGAATGAAGTAACTGAATTTGCAAAAACATGTGTTGCTGATGAGTCAGCTGAAAATTGTGACAAATCACTTCATACCCTTTTTGGAGACAAATTATGCACAGTTGCAACTCTTCGTGAAACCTATGGTGAAATGGCTGACTGCTGTGCAAAACAAGAACCTGAGAGAAATGAATGCTTCTTGCAACACAAAGATGACAACCCAAACCTCCCCCGATTGGTGAGACCAGAGGTTGATGTGATGTGCACTGCTTTTCATGACAATGAAGAGACATTTTTGAAAAAATACTTATATGAAATTGCCAGAAGACATCCTTACTTTTATGCCCCGGAACTCCTTTTCTTTGCTAAAAGGTATAAAGCTGCTTTTACAGAATGTTGCCAAGCTGCTGATAAAGCTGCCTGCCTGTTGCCAAAGCTCGATGAACTTCGGGATGAAGGGAAGGCTTCGTCTGCCAAACAGAGACTCAAGTGTGCCAGTCTCCAAAAATTTGGAGAAAGAGCTTTCAAAGCATGGGCAGTAGCTCGCCTGAGCCAGAGATTTCCCAAAGCTGAGTTTGCAGAAGTTTCCAAGTTAGTGACAGATCTTACCAAAGTCCACACGGAATGCTGCCATGGAGATCTGCTTGAATGTGCTGATGACAGGGCGGACCTTGCCAAGTATATCTGTGAAAATCAAGATTCGATCTCCAGTAAACTGAAGGAATGCTGTGAAAAACCTCTGTTGGAAAAATCCCACTGCATTGCCGAAGTGGAAAATGATGAGATGCCTGCTGACTTGCCTTCATTAGCTGCTGATTTTGTTGAAAGTAAGGATGTTTGCAAAAACTATGCTGAGGCAAAGGATGTCTTCCTGGGCATGTTTTTGTATGAATATGCAAGAAGGCATCCTGATTACTCTGTCGTGCTGCTGCTGAGACTTGCCAAGACATATGAAACCACTCTAGAGAAGTGCTGTGCCGCTGCAGATCCTCATGAATGCTATGCCAAAGTGTTCGATGAATTTAAACCTCTTGTGGAAGAGCCTCAGAATTTAATCAAACAAAATTGTGAGCTTTTTGAGCAGCTTGGAGAGTACAAATTCCAGAATGCGCTATTAGTTCGTTACACCAAGAAAGTACCCCAAGTGTCAACTCCAACTCTTGTAGAGGTCTCAAGAAACCTAGGAAAAGTGGGCAGCAAATGTTGTAAACATCCTGAAGCAAAAAGAATGCCCTGTGCAGAAGACTATCTATCCGTGGTCCTGAACCAGTTATGTGTGTTGCATGAGAAAACGCCAGTAAGTGACAGAGTCACCAAATGCTGCACAGAATCCTTGGTGAACAGGCGACCATGCTTTTCAGCTCTGGAAGTCGATGAAACATACGTTCCCAAAGAGTTTAATGCTGAAACATTCACCTTCCATGCAGATATATGCACACTTTCTGAGAAGGAGAGACAAATCAAGAAACAAACTGCACTTGTTGAGCTTGTGAAACACAAGCCCAAGGCAACAAAAGAGCAACTGAAAGCTGTTATGGATGATTTCGCAGCTTTTGTAGAGAAGTGCTGCAAGGCTGACGATAAGGAGACCTGCTTTGCCGAGGAGGGTAAAAAAACTTGTTGCTGCAAGTCAAGCTGCCTTAGGCTTATAACATCACATTTAAAAGCATCTCAGCCTACCATGAGAATAAGAGAAAGAAAATGAAGATCAAAAGCTTATTCATCTGTTTTTCTTTTTCGTTGGTGTAAAGCCAACACCCTGTCTAAAAAACATAAATTTCTTTAATCATTTTGCCTCTTTTCTCTGTGCTTCAATTAATAAAAAATGGAAAGAATCT start: 7 end: 2056
Align Title:gi|23243417|gb|BC036003.1| Homo sapiens albumin, mRNA (cDNA clone MGC:32850 IMAGE:4724105), complete cds
Align Len: 2087
Score: 4052.0 Bits: 3654.91 E-val: 0.0
N.aligns:None Ident:2041 Pos.:2041 Gaps:0 Align len:2051
Strand: (None, None) Frame: (1, 1)
Query: TCTCTTCTGTCAACCCCACGCGCCTTTGGCACAATGAAGTGGGTAACCTTTATTTCCCTTCTTTTTCTCTTTAGCTCGGCTTATTCCAGGGGTGTGTTTCGTCGAGATGCACACAAGAGTGAGGTTGCTCATCGGTTTAAAGATTTGGGAGAAGAAAATTTCAAAGCCTTAGTGTTGATTGCCTTTGCTCAGTATCTTCAGCAGTGTCCATTTGAAGATCATGTAAAATTAGTGAATGAAGTAACTGAATTTGCAAAAACATGTGTTGCTGATGAGTCAGCTGAAAATTGTGACAAATCACTTCATACCCTTTTTGGAGACAAATTATGCACAGTTGCAACTCTTCGTGAAACCTATGGTGAAATGGCTGACTGCTGTGCAAAACAAGAACCTGAGAGAAATGAATGCTTCTTGCAACACAAAGATGACAACCCAAACCTCCCCCGATTGGTGAGACCAGAGGTTGATGTGATGTGCACTGCTTTTCATGACAATGAAGAGACATTTTTGAAAAAATACTTATATGAAATTGCCAGAAGACATCCTTACTTCTATGCCCCGGAACTCCTTTTCTTTGCTAAAAGGTATAAAGCTGCTTTTACAGAATGTTGCCAAGCTGCTGATAAGGCTGCCTGCCTGTTGCCAAAGCTCGATGAACTTCGGGATGAAGGGAAGGCTTCGTCTGCCAAACAGAGGCTCAAGTGTGCCAGTCTCCAAAAATTTGGAGAAAGAGCTTTCAAAGCATGGGCAGTAGCTCGCCTGAGCCAGAGATTTCCCAAAGCTGAGTTTGCAGAAGTTTCCAAGTTAGTGACAGATCTTACCAAAGTCCACACGGAATGCTGCCATGGAGATCTGCTTGAATGTGCTGATGACAGGGCGGACCTTGCCAAGTATATCTGTGAAAATCAAGATTCGATCTCCAGTAAACTGAAGGAATGCTGTGAAAAACCTCTGTTGGAAAAATCCCACTGCATTGCCGAAGTGGAAAATGATGAGATGCCTGCTGACTTGCCTTCATTAGCTGCTGATTTTGTTGAAAGTAAGGATGTTTGCAAAAACTATGCTGAGGCAAAGGATGTCTTCCTGGGCATGTTTTTGTATGAATATGCAAGAAGGCATCCTGATTACTCTGTCGTGCTGCTGCTGAGACTTGCCAAGACATATAAAACCACTCTAGAGAAGTGCTGTGCCGCTGCAGATCCTCATGAATGCTATGCCAAAGTGTTCGATGAATTTAAACCTCTTGTGGAAGAGCCTCAGAATTTAATCAAACAAAATTGTGAGCTTTTTGAGCAGCTTGGAGAGTACAAATTCCAGAATGCGCTATTAGTTCGTTACACCAAGAAAGTACCCCAAGTGTCAACTCCAACTCTTGTAGAGGTCTCAAGAAACCTAGGAAAAGTGGGCAGCAAATGTTGTAAACATCCTGAAGCAAAAAGAATGCCCTGTGCAGAAGACTATCTATCCGTGGTCCTGAACCAGTTATGTGTGTTGCATGAGAAAACGCCAGTAAGTGACAGAGTCACCAAATGCTGCACAGAATCCTTGGTGAACAGGCGACCATGCTTTTCAGCTCTGGAAGTCGATGAAACATACGTTCCCAAAGAGTTTAATGCTGAAACATTCACCTTCCATGCAGATATATGCACACTTTCTGAGAAGGAGAGACAAATCAAGAAACAAACTGCACTTGTTGAGCTTGTGAAACACAAGCCCAAGGCAACAAAAGAGCAACTGAAAGCTGTTATGGATGATTTCGCAGCTTTTGTAGAGAAGTGCTGCAAGGCTGACGATAAGGAGACCTGCTTTGCCGAGGAGGGTAAAAAACTTGTTGCTGCAAGTCGAGCTGCCTTAGGCTTATAACATCACATTTAAAAGCATCTCAGCCTACCATGAGAATAAGAGAAAGAAAATGAAGATCAAAAGCTTATTCATCTGTTTTTCTTTTTCGTTGGTGTAAAGCCAACACCCTGTCTAAAAAACATAAATTTCTTTAATCATTTTGCCTCTTTTCTCTGTGCTTCAATTAATAAAAAATGGAAAGAATCTAA start: 1 end: 2051
Match: ||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| ||
Subjc: TCTCTTCTGTCAACCCCACACGCCTTTGGCACAATGAAGTGGGTAACCTTTATTTCCCTTCTTTTTCTCTTTAGCTCGGCTTATTCCAGGGGTGTGTTTCGTCGAGATGCACACAAGAGTGAGGTTGCTCATCGGTTTAAAGATTTGGGAGAAGAAAATTTCAAAGCCTTGGTGTTGATTGCCTTTGCTCAGTATCTTCAGCAGTGTCCATTTGAAGATCATGTAAAATTAGTGAATGAAGTAACTGAATTTGCAAAAACATGTGTTGCTGATGAGTCAGCTGAAAATTGTGACAAATCACTTCATACCCTTTTTGGAGACAAATTATGCACAGTTGCAACTCTTCGTGAAACCTATGGTGAAATGGCTGACTGCTGTGCAAAACAAGAACCTGAGAGAAATGAATGCTTCTTGCAACACAAAGATGACAACCCAAACCTCCCCCGATTGGTGAGACCAGAGGTTGATGTGATGTGCACTGCTTTTCATGACAATGAAGAGACATTTTTGAAAAAATACTTATATGAAATTGCCAGAAGACATCCTTACTTTTATGCCCCGGAACTCCTTTTCTTTGCTAAAAGGTATAAAGCTGCTTTTACAGAATGTTGCCAAGCTGCTGATAAAGCTGCCTGCCTGTTGCCAAAGCTCGATGAACTTCGGGATGAAGGGAAGGCTTCGTCTGCCAAACAGAGACTCAAGTGTGCCAGTCTCCAAAAATTTGGAGAAAGAGCTTTCAAAGCATGGGCAGTAGCTCGCCTGAGCCAGAGATTTCCCAAAGCTGAGTTTGCAGAAGTTTCCAAGTTAGTGACAGATCTTACCAAAGTCCACACGGAATGCTGCCATGGAGATCTGCTTGAATGTGCTGATGACAGGGCGGACCTTGCCAAGTATATCTGTGAAAATCAAGATTCGATCTCCAGTAAACTGAAGGAATGCTGTGAAAAACCTCTGTTGGAAAAATCCCACTGCATTGCCGAAGTGGAAAATGATGAGATGCCTGCTGACTTGCCTTCATTAGCTGCTGATTTTGTTGAAAGTAAGGATGTTTGCAAAAACTATGCTGAGGCAAAGGATGTCTTCCTGGGCATGTTTTTGTATGAATATGCAAGAAGGCATCCTGATTACTCTGTCGTGCTGCTGCTGAGACTTGCCAAGACATATGAAACCACTCTAGAGAAGTGCTGTGCCGCTGCAGATCCTCATGAATGCTATGCCAAAGTGTTCGATGAATTTAAACCTCTTGTGGAAGAGCCTCAGAATTTAATCAAACAAAATTGTGAGCTTTTTGAGCAGCTTGGAGAGTACAAATTCCAGAATGCGCTATTAGTTCGTTACACCAAGAAAGTACCCCAAGTGTCAACTCCAACTCTTGTAGAGGTCTCAAGAAACCTAGGAAAAGTGGGCAGCAAATGTTGTAAACATCCTGAAGCAAAAAGAATGCCCTGTGCAGAAGACTATCTATCCGTGGTCCTGAACCAGTTATGTGTGTTGCATGAGAAAACGCCAGTAAGTGACAGAGTCACCAAATGCTGCACAGAATCCTTGGTGAACAGGCGTCCATGCTTTTCAGCTCTGGAAGTCGATGAAACATACGTTCCCAAAGAGTTTAATGCTGAAACATTCACCTTCCATGCAGATATATGCACACTTTCTGAGAAGGAGAGACAAATCAAGAAACAAACTGCACTTGTTGAGCTCGTGAAACACAAGCCCAAGGCAACAAAAGAGCAACTGAAAGCTGTTATGGATGATTTCGCAGCTTTTGTAGAGAAGTGCTGCAAGGCTGACGATAAGGAGACCTGCTTTGCCGAGGAGGGTAAAAAACTTGTTGCTGCAAGTCAAGCTGCCTTAGGCTTATAACATCACATTTAAAAGCATCTCAGCCTACCATGAGAATAAGAGAAAGAAAATGAAGATCAAAAGCTTATTCATCTGTTTTTCTTTTTCGTTGGTGTAAAGCCAACACCCTGTCTAAAAAACATAAATTTCTTTAATCATTTTGCCTCTTTTCTCTGTGCTTCAATTAATAAAAAATGGAAAGAATCGAA start: 9 end: 2059
Align Title:gi|158258946|dbj|AK292755.1| Homo sapiens cDNA FLJ78413 complete cds, highly similar to Homo sapiens albumin, mRNA
Align Len: 2054
Score: 4036.0 Bits: 3640.48 E-val: 0.0
N.aligns:None Ident:2037 Pos.:2037 Gaps:3 Align len:2049
Strand: (None, None) Frame: (1, 1)
Query: TCTCTTCTGTCAACCCCACGCGCCTTTGGCACAATGAAGTGGGTAACCTTTATTTCCCTTCTTTTTCTCTTTAGCTCGGCTTATTCCAGGGGTGTGTTTCGTCGAGATGCACACAAGAGTGAGGTTGCTCATCGGTTTAAAGATTTGGGAGAAGAAAATTTCAAAGCCTTAGTGTTGATTGCCTTTGCTCAGTATCTTCAGCAGTGTCCATTTGAAGATCATGTAAAATTAGTGAATGAAGTAACTGAATTTGCAAAAACATGTGTTGCTGATGAGTCAGCTGAAAATTGTGACAAATCACTTCATACCCTTTTTGGAGACAAATTATGCACAGTTGCAACTCTTCGTGAAACCTATGGTGAAATGGCTGACTGCTGTGCAAAACAAGAACCTGAGAGAAATGAATGCTTCTTGCAACACAAAGATGACAACCCAAACCTCCCCCGATTGGTGAGACCAGAGGTTGATGTGATGTGCACTGCTTTTCATGACAATGAAGAGACATTTTTGAAAAAATACTTATATGAAATTGCCAGAAGACATCCTTACTTCTATGCCCCGGAACTCCTTTTCTTTGCTAAAAGGTATAAAGCTGCTTTTACAGAATGTTGCCAAGCTGCTGATAAGGCTGCCTGCCTGTTGCCAAAGCTCGATGAACTTCGGGATGAAGGGAAGGCTTCGTCTGCCAAACAGAGGCTCAAGTGTGCCAGTCTCCAAAAATTTGGAGAAAGAGCTTTCAAAGCATGGGCAGTAGCTCGCCTGAGCCAGAGATTTCCCAAAGCTGAGTTTGCAGAAGTTTCCAAGTTAGTGACAGATCTTACCAAAGTCCACACGGAATGCTGCCATGGAGATCTGCTTGAATGTGCTGATGACAGGGCGGACCTTGCCAAGTATATCTGTGAAAATCAAGATTCGATCTCCAGTAAACTGAAGGAATGCTGTGAAAAACCTCTGTTGGAAAAATCCCACTGCATTGCCGAAGTGGAAAATGATGAGATGCCTGCTGACTTGCCTTCATTAGCTGCTGATTTTGTTGAAAGTAAGGATGTTTGCAAAAACTATGCTGAGGCAAAGGATGTCTTCCTGGGCATGTTTTTGTATGAATATGCAAGAAGGCATCCTGATTACTCTGTCGTGCTGCTGCTGAGACTTGCCAAGACATATAAAACCACTCTAGAGAAGTGCTGTGCCGCTGCAGATCCTCATGAATGCTATGCCAAAGTGTTCGATGAATTTAAACCTCTTGTGGAAGAGCCTCAGAATTTAATCAAACAAAATTGTGAGCTTTTTGAGCAGCTTGGAGAGTACAAATTCCAGAATGCGCTATTAGTTCGTTACACCAAGAAAGTACCCCAAGTGTCAACTCCAACTCTTGTAGAGGTCTCAAGAAACCTAGGAAAAGTGGGCAGCAAATGTTGTAAACATCCTGAAGCAAAAAGAATGCCCTGTGCAGAAGACTATCTATCCGTGGTCCTGAACCAGTTATGTGTGTTGCATGAGAAAACGCCAGTAAGTGACAGAGTCACCAAATGCTGCACAGAATCCTTGGTGAACAGGCGACCATGCTTTTCAGCTCTGGAAGTCGATGAAACATACGTTCCCAAAGAGTTTAATGCTGAAACATTCACCTTCCATGCAGATATATGCACACTTTCTGAGAAGGAGAGACAAATCAAGAAACAAACTGCACTTGTTGAGCTTGTGAAACACAAGCCCAAGGCAACAAAAGAGCAACTGAAAGCTGTTATGGATGATTTCGCAGCTTTTGTAGAGAAGTGCTGCAAGGCTGACGATAAGGAGACCTGCTTTGCCGAGGAGGGTAAAAAACTTGTTGCTGCAAGTCGAGCTGCCTTAGGCTTATAACATCACATTTAAAAGCATCTCAGCCTACCATGAGAATAAGAGAAAGAAAATGAAGATCAAAAGCTTATTCATCTGTTTTTCTTTTTCGTTGGTGTAAAGCCAACACCCTGTCTAAAAAACATAAATTTCTTTAATCATTTTGCCTCTTTTCTCTGTGCTTCAATTAATAAAAAATGGAAAGAATCT start: 1 end: 2049
Match: ||||||||||||||||||| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| ||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Subjc: TCTCTTCTGTCAACCCCACACGCCTTTGGCACAATGAAGTGGGTAACCTTTATTTCCCTTCTTTTTCTCTTTAGCTCGGCTTATTCCAGGGGTGTGTTTCGTCGAGATGCACACAAGAGTGAGGTTGCTCATCGGTTTAAAGATTTGGGAGAA---AATTTCAAAGCCTTGGTGTTGATTGCCTTTGCTCAGTATCTTCAGCAGTGTCCATTTGAAGATCATGTAAAATTAGTGAATGAAGTAACTGAATTTGCAAAAACATGTGTTGCTGATGAGTCAGCTGAAAATTGTGACAAATCACTTCATACCCTTTTTGGAGACAAATTATGCACAGTTGCAACTCTTCGTGAAACCTATGGTGAAATGGCTGACTGCTGTGCAAAACAAGAACCTGAGAGAAATGAATGCTTCTTGCAACACAAAGATGACAACCCAAACCTCCCCCGATTGGTGAGACCAGAGGTTGATGTGATGTGCACTGCTTTTCATGACAATGAAGAGACATTTTTGAAAAAATACTTATATGAAATTGCCAGAAGACATCCTTACTTTTATGCCCCGGAACTCCTTTTCTTTGCTAAAAGGTATAAAGCTGCTTTTACAGAATGTTGCCAAGCTGCTGATAAAGCTGCCTGCCTGTTGCCAAAGCTCGATGAACTTTGGGATGAAGGGAAGGCTTCGTCTGCCAAACAGAGACTCAAGTGTGCCAGTCTCCAAAAATTTGGAGAAAGAGCTTTCAAAGCATGGGCAGTAGCTCGCCTGAGCCAGAGATTTCCCAAAGCTGAGTTTGCAGAAGTTTCCAAGTTAGTGACAGATCTTACCAAAGTCCACACGGAATGCTGCCATGGAGATCTGCTTGAATGTGCTGATGACAGGGCGGACCTTGCCAAGTATATCTGTGAAAATCAAGATTCGATCTCCAGTAAACTGAAGGAATGCTGTGAAAAACCTCTGTTGGAAAAATCCCACTGCATTGCCGAAGTGGAAAATGATGAGATGCCTGCTGACTTGCCTTCATTAGCTGCTGATTTTGTTGAAAGTAAGGATGTTTGCAAAAACTATGCTGAGGCAAAGGATGTCTTCCTGGGCATGTTTTTGTATGAATATGCAAGAAGGCATCCTGATTACTCTGTCGTGCTGCTGCTGAGACTTGCCAAGACATATGAAACCACTCTAGAGAAGTGCTGTGCCGCTGCAGATCCTCATGAATGCTATGCCAAAGTGTTCGATGAATTTAAACCTCTTGTGGAAGAGCCTCAGAATTTAATCAAACAAAATTGTGAGCTTTTTGAGCAGCTTGGAGAGTACAAATTCCAGAATGCGCTATTAGTTCGTTACACCAAGAAAGTACCCCAAGTGTCAACTCCAACTCTTGTAGAGGTCTCAAGAAACCTAGGAAAAGTGGGCAGCAAATGTTGTAAACATCCTGAAGCAAAAAGAATGCCCTGTGCAGAAGACTATCTATCCGTGGTCCTGAACCAGTTATGTGTGTTGCATGAGAAAACGCCAGTAAGTGACAGAGTCACCAAATGCTGCACAGAATCCTTGGTGAACAGGCGACCATGCTTTTCAGCTCTGGAAGTCGATGAAACATACGTTCCCAAAGAGTTTAATGCTGAAACATTCACCTTCCATGCAGATATATGCACACTTTCTGAGAAGGAGAGACAAATCAAGAAACAAACTGCACTTGTTGAGCTCGTGAAACACAAGCCCAAGGCAACAAAAGAGCAACTGAAAGCTGTTATGGATGATTTCGCAGCTTTTGTAGAGAAGTGCTGCAAGGCTGACGATAAGGAGACCTGCTTTGCCGAGGAGGGTAAAAAACTTGTTGCTGCAAGTCAAGCTGCCTTAGGCTTATAACATCACATTTAAAAGCATCTCAGCCTACCATGAGAATAAGAGAAAGAAAATGAAGATCAAAAGCTTATTCATCTGTTTTTCTTTTTCGTTGGTGTAAAGCCAACACCCTGTCTAAAAAACATAAATTTCTTTAATCATTTTGCCTCTTTTCTCTGTGCTTCAATTAATAAAAAATGGAAAGAATCT start: 9 end: 2054
Align Title:gi|28589|emb|V00494.1| Human messenger RNA for serum albumin (HSA)
Align Len: 2055
Score: 4033.0 Bits: 3637.78 E-val: 0.0
N.aligns:None Ident:2036 Pos.:2036 Gaps:0 Align len:2049
Strand: (None, None) Frame: (1, 1)
Query: TCTCTTCTGTCAACCCCACGCGCCTTTGGCACAATGAAGTGGGTAACCTTTATTTCCCTTCTTTTTCTCTTTAGCTCGGCTTATTCCAGGGGTGTGTTTCGTCGAGATGCACACAAGAGTGAGGTTGCTCATCGGTTTAAAGATTTGGGAGAAGAAAATTTCAAAGCCTTAGTGTTGATTGCCTTTGCTCAGTATCTTCAGCAGTGTCCATTTGAAGATCATGTAAAATTAGTGAATGAAGTAACTGAATTTGCAAAAACATGTGTTGCTGATGAGTCAGCTGAAAATTGTGACAAATCACTTCATACCCTTTTTGGAGACAAATTATGCACAGTTGCAACTCTTCGTGAAACCTATGGTGAAATGGCTGACTGCTGTGCAAAACAAGAACCTGAGAGAAATGAATGCTTCTTGCAACACAAAGATGACAACCCAAACCTCCCCCGATTGGTGAGACCAGAGGTTGATGTGATGTGCACTGCTTTTCATGACAATGAAGAGACATTTTTGAAAAAATACTTATATGAAATTGCCAGAAGACATCCTTACTTCTATGCCCCGGAACTCCTTTTCTTTGCTAAAAGGTATAAAGCTGCTTTTACAGAATGTTGCCAAGCTGCTGATAAGGCTGCCTGCCTGTTGCCAAAGCTCGATGAACTTCGGGATGAAGGGAAGGCTTCGTCTGCCAAACAGAGGCTCAAGTGTGCCAGTCTCCAAAAATTTGGAGAAAGAGCTTTCAAAGCATGGGCAGTAGCTCGCCTGAGCCAGAGATTTCCCAAAGCTGAGTTTGCAGAAGTTTCCAAGTTAGTGACAGATCTTACCAAAGTCCACACGGAATGCTGCCATGGAGATCTGCTTGAATGTGCTGATGACAGGGCGGACCTTGCCAAGTATATCTGTGAAAATCAAGATTCGATCTCCAGTAAACTGAAGGAATGCTGTGAAAAACCTCTGTTGGAAAAATCCCACTGCATTGCCGAAGTGGAAAATGATGAGATGCCTGCTGACTTGCCTTCATTAGCTGCTGATTTTGTTGAAAGTAAGGATGTTTGCAAAAACTATGCTGAGGCAAAGGATGTCTTCCTGGGCATGTTTTTGTATGAATATGCAAGAAGGCATCCTGATTACTCTGTCGTGCTGCTGCTGAGACTTGCCAAGACATATAAAACCACTCTAGAGAAGTGCTGTGCCGCTGCAGATCCTCATGAATGCTATGCCAAAGTGTTCGATGAATTTAAACCTCTTGTGGAAGAGCCTCAGAATTTAATCAAACAAAATTGTGAGCTTTTTGAGCAGCTTGGAGAGTACAAATTCCAGAATGCGCTATTAGTTCGTTACACCAAGAAAGTACCCCAAGTGTCAACTCCAACTCTTGTAGAGGTCTCAAGAAACCTAGGAAAAGTGGGCAGCAAATGTTGTAAACATCCTGAAGCAAAAAGAATGCCCTGTGCAGAAGACTATCTATCCGTGGTCCTGAACCAGTTATGTGTGTTGCATGAGAAAACGCCAGTAAGTGACAGAGTCACCAAATGCTGCACAGAATCCTTGGTGAACAGGCGACCATGCTTTTCAGCTCTGGAAGTCGATGAAACATACGTTCCCAAAGAGTTTAATGCTGAAACATTCACCTTCCATGCAGATATATGCACACTTTCTGAGAAGGAGAGACAAATCAAGAAACAAACTGCACTTGTTGAGCTTGTGAAACACAAGCCCAAGGCAACAAAAGAGCAACTGAAAGCTGTTATGGATGATTTCGCAGCTTTTGTAGAGAAGTGCTGCAAGGCTGACGATAAGGAGACCTGCTTTGCCGAGGAGGGTAAAAAACTTGTTGCTGCAAGTCGAGCTGCCTTAGGCTTATAACATCACATTTAAAAGCATCTCAGCCTACCATGAGAATAAGAGAAAGAAAATGAAGATCAAAAGCTTATTCATCTGTTTTTCTTTTTCGTTGGTGTAAAGCCAACACCCTGTCTAAAAAACATAAATTTCTTTAATCATTTTGCCTCTTTTCTCTGTGCTTCAATTAATAAAAAATGGAAAGAATCT start: 1 end: 2049
Match: ||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| ||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Subjc: TCTCTTCTGTCAACCCCACACGCCTTTGGCACAATGAAGTGGGTAACCTTTATTTCCCTTCTTTTTCTCTTTAGCTCGGCTTATTCCAGGGGTGTGTTTCGTCGAGATGCACACAAGAGTGAGGTTGCTCATCGGTTTAAAGATTTGGGAGAAGAAAATTTCAAAGCCTTGGTGTTGATTGCCTTTGCTCAGTATCTTCAGCAGTGTCCATTTGAAGATCATGTAAAATTAGTGAATGAAGTAACTGAATTTGCAAAAACATGTGTTGCTGATGAGTCAGCTGAAAATTGTGACAAATCACTTCATACCCTTTTTGGAGACAAATTATGCACAGTTGCAACTCTTCGTGAAACCTATGGTGAAATGGCTGACTGCTGTGCAAAACAAGAACCTGGGAGAAATGAATGCTTCTTGCAACACAAAGATGACAACCCAAACCTCCCCCGATTGGTGAGACCAGAGGTTGATGTGATGTGCACTGCTTTTCATGACAATGAAGAGACATTTTTGAAAAAATACTTATATGAAATTGCCAGAAGACATCCTTACTTTTATGCCCCGGAACTCCTTTTCTTTGCTAAAAGGTATAAAGCTGCTTTTACAGAATGTTGCCAAGCTGCTGATAAAGCTGCCTGCCTGTTGCCAAAGCTCGATGAACTTCGGGATGAAGGGAAGGCTTCGTCTGCCAAACAGAGACTCAAGTGTGCCAGTCTCCAAAAATTTGGAGAAAGAGCTTTCAAAGCATGGGCAGTAGCTCGCCTGAGCCAGAGATTTCCCAAAGCTGAGTTTGCAGAAGTTTCCAAGTTAGTGACAGATCTTACCAAAGTCCACACGGAATGCTGCCATGGAGATCTGCTTGAATGTGCTGATGACAGGGCGGACCTTGCCAAGTATATCTGTGAAAATCAAGATTCGATCTCCAGTAAACTGAAGGAATGCTGTGAAAAACCTCTGTTGGAAAAATCCCACTGCATTGCCGAAGTGGAAAATGATGAGATGCCTGCTGACTTGCCTTCATTAGCTGCTGATTTTGTTGAAAGTAAGGATGTTTGCAAAAACTATGCTGAGGCAAAGGATGTCTTCTTGGGCATGTTTTTGTATGAATATGCAAGAAGGCATCCTGATTACTCTGTCGTGCTGCTGCTGAGACTTGCCAAGACATATGAAACCACTCTAGAGAAGTGCTGTGCCGCTGCAGATCCTCATGAATGCTATGCCAAAGTGTTCGATGAATTTAAACCTCTTGTGGAAGAGCCTCAGAATTTAATCAAACAAAATTGTGAGCTTTTTGAGCAGCTTGGAGAGTACAAATTCCAGAATGCGCTGTTAGTTCGTTACACCAAGAAAGTACCCGAAGTGTCAACTCCAACTCTTGTAGAGGTCTCAAGAAACCTAGGAAAAGTGGGCAGCAAATGTTGTAAACATCCTGAAGCAAAAAGAATGCCCTGTGCAGAAGACTATCTATCCGTGGTCCTGAACCAGTTATGTGTGTTGCATGAGAAAACGCCAGTAAGTGACAGAGTCACCAAATGCTGCACAGAATCCTTGGTGAACAGGCGACCATGCTTTTCAGCTCTGGAAGTCGATGAAACATACGTTCCCAAAGAGTTTAATGCTGAAACATTCACCTTCCATGCAGATATATGCACACTTTCTGAGAAGGAGAGACAAATCAAGAAACAAACTGCACTTGTTGAGCTCGTGAAACACAAGCCCAAGGCAACAAAAGAGCAACTGAAAGCTGTTATGGATGATTTCGCTGCTTTTGTAGAGAAGTGCTGCAAGGCTGACGATAAGGAGACCTGCTTTGCCGAGGAGGGTAAAAAACTTGTTGCTGCAAGTCAAGCTGCCTTAGGCTTATAACATCACATTTAAAAGCATCTCAGCCTACCATGAGAATAAGAGAAAGAAAATGAAGATCAAAAGCTTATTCATCTGTTTTTCTTTTTCGTTGGTGTAAAGCCAACACCCTGTCTAAAAAACATAAATTTCTTTAATCATTTTGCCTCTTTTCTCTGTGCTTCAATTAATAAAAAATGGAAAGAATCT start: 7 end: 2055
Align Title:gi|21706455|gb|BC034023.1| Homo sapiens albumin, mRNA (cDNA clone MGC:22784 IMAGE:4734617), complete cds
Align Len: 2068
Score: 4029.0 Bits: 3634.17 E-val: 0.0
N.aligns:None Ident:2028 Pos.:2028 Gaps:0 Align len:2037
Strand: (None, None) Frame: (1, 1)
Query: TCTCTTCTGTCAACCCCACGCGCCTTTGGCACAATGAAGTGGGTAACCTTTATTTCCCTTCTTTTTCTCTTTAGCTCGGCTTATTCCAGGGGTGTGTTTCGTCGAGATGCACACAAGAGTGAGGTTGCTCATCGGTTTAAAGATTTGGGAGAAGAAAATTTCAAAGCCTTAGTGTTGATTGCCTTTGCTCAGTATCTTCAGCAGTGTCCATTTGAAGATCATGTAAAATTAGTGAATGAAGTAACTGAATTTGCAAAAACATGTGTTGCTGATGAGTCAGCTGAAAATTGTGACAAATCACTTCATACCCTTTTTGGAGACAAATTATGCACAGTTGCAACTCTTCGTGAAACCTATGGTGAAATGGCTGACTGCTGTGCAAAACAAGAACCTGAGAGAAATGAATGCTTCTTGCAACACAAAGATGACAACCCAAACCTCCCCCGATTGGTGAGACCAGAGGTTGATGTGATGTGCACTGCTTTTCATGACAATGAAGAGACATTTTTGAAAAAATACTTATATGAAATTGCCAGAAGACATCCTTACTTCTATGCCCCGGAACTCCTTTTCTTTGCTAAAAGGTATAAAGCTGCTTTTACAGAATGTTGCCAAGCTGCTGATAAGGCTGCCTGCCTGTTGCCAAAGCTCGATGAACTTCGGGATGAAGGGAAGGCTTCGTCTGCCAAACAGAGGCTCAAGTGTGCCAGTCTCCAAAAATTTGGAGAAAGAGCTTTCAAAGCATGGGCAGTAGCTCGCCTGAGCCAGAGATTTCCCAAAGCTGAGTTTGCAGAAGTTTCCAAGTTAGTGACAGATCTTACCAAAGTCCACACGGAATGCTGCCATGGAGATCTGCTTGAATGTGCTGATGACAGGGCGGACCTTGCCAAGTATATCTGTGAAAATCAAGATTCGATCTCCAGTAAACTGAAGGAATGCTGTGAAAAACCTCTGTTGGAAAAATCCCACTGCATTGCCGAAGTGGAAAATGATGAGATGCCTGCTGACTTGCCTTCATTAGCTGCTGATTTTGTTGAAAGTAAGGATGTTTGCAAAAACTATGCTGAGGCAAAGGATGTCTTCCTGGGCATGTTTTTGTATGAATATGCAAGAAGGCATCCTGATTACTCTGTCGTGCTGCTGCTGAGACTTGCCAAGACATATAAAACCACTCTAGAGAAGTGCTGTGCCGCTGCAGATCCTCATGAATGCTATGCCAAAGTGTTCGATGAATTTAAACCTCTTGTGGAAGAGCCTCAGAATTTAATCAAACAAAATTGTGAGCTTTTTGAGCAGCTTGGAGAGTACAAATTCCAGAATGCGCTATTAGTTCGTTACACCAAGAAAGTACCCCAAGTGTCAACTCCAACTCTTGTAGAGGTCTCAAGAAACCTAGGAAAAGTGGGCAGCAAATGTTGTAAACATCCTGAAGCAAAAAGAATGCCCTGTGCAGAAGACTATCTATCCGTGGTCCTGAACCAGTTATGTGTGTTGCATGAGAAAACGCCAGTAAGTGACAGAGTCACCAAATGCTGCACAGAATCCTTGGTGAACAGGCGACCATGCTTTTCAGCTCTGGAAGTCGATGAAACATACGTTCCCAAAGAGTTTAATGCTGAAACATTCACCTTCCATGCAGATATATGCACACTTTCTGAGAAGGAGAGACAAATCAAGAAACAAACTGCACTTGTTGAGCTTGTGAAACACAAGCCCAAGGCAACAAAAGAGCAACTGAAAGCTGTTATGGATGATTTCGCAGCTTTTGTAGAGAAGTGCTGCAAGGCTGACGATAAGGAGACCTGCTTTGCCGAGGAGGGTAAAAAACTTGTTGCTGCAAGTCGAGCTGCCTTAGGCTTATAACATCACATTTAAAAGCATCTCAGCCTACCATGAGAATAAGAGAAAGAAAATGAAGATCAAAAGCTTATTCATCTGTTTTTCTTTTTCGTTGGTGTAAAGCCAACACCCTGTCTAAAAAACATAAATTTCTTTAATCATTTTGCCTCTTTTCTCTGTGCTTCAATTAATAAAAAA start: 1 end: 2037
Match: ||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| ||||||
Subjc: TCTCTTCTGTCAACCCCACACGCCTTTGGCACAATGAAGTGGGTAACCTTTATTTCCCTTCTTTTTCTCTTTAGCTCGGCTTATTCCAGGGGTGTGTTTCGTCGAGATGCACACAAGAGTGAGGTTGCTCATCGGTTTAAAGATTTGGGAGAAGAAAATTTCAAAGCCTTGGTGTTGATTGCCTTTGCTCAGTATCTTCAGCAGTGTCCATTTGAAGATCATGTAAAATTAGTGAATGAAGTAACTGAATTTGCAAAAACATGTGTTGCTGATGAGTCAGCTGAAAATTGTGACAAATCACTTCATACCCTTTTTGGAGACAAATTATGCACAGTTGCAACTCTTCGTGAAACCTATGGTGAAATGGCTGACTGCTGTGCAAAACAAGAACCTGAGAGAAATGAATGCTTCTTGCAACACAAAGATGACAACCCAAACCTCCCCCGATTGGTGAGACCAGAGGTTGATGTGATGTGCACTGCTTTTCATGACAATGAAGAGACATTTTTGAAAAAATACTTATATGAAATTGCCAGAAGACATCCTTACTTTTATGCCCCGGAACTCCTTTTCTTTGCTAAAAGGTATAAAGCTGCTTTTACAGAATGTTGCCAAGCTGCTGATAAAGCTGCCTGCCTGTTGCCAAAGCTCGATGAACTTCGGGATGAAGGGAAGGCTTCGTCTGCCAAACAGAGACTCAAGTGTGCCAGTCTCCAAAAATTTGGAGAAAGAGCTTTCAAAGCATGGGCAGTAGCTCGCCTGAGCCAGAGATTTCCCAAAGCTGAGTTTGCAGAAGTTTCCAAGTTAGTGACAGATCTTACCAAAGTCCACACGGAATGCTGCCATGGAGATCTGCTTGAATGTGCTGATGACAGGGCGGACCTTGCCAAGTATATCTGTGAAAATCAAGATTCGATCTCCAGTAAACTGAAGGAATGCTGTGAAAAACCTCTGTTGGAAAAATCCCACTGCATTGCCGAAGTGGAAAATGATGAGATGCCTGCTGACTTGCCTTCATTAGCTGCTGATTTTGTTGAAAGTAAGGATGTTTGCAAAAACTATGCTGAGGCAAAGGATGTCTTCCTGGGCATGTTTTTGTATGAATATGCAAGAAGGCATCCTGATTACTCTGTCGTGCTGCTGCTGAGACTTGCCAAGACATATGAAACCACTCTAGAGAAGTGCTGTGCCGCTGCAGATCCTCATGAATGCTATGCCAAAGTGTTCGATGAATTTAAACCTCTTGTGGAAGAGCCTCAGAATTTAATCAAACAAAATTGTGAGCTTTTTGAGCAGCTTGGAGAGTACAAATTCCAGAATGCGCTATTAGTTCGTTACACCAAGAAAGTACCCCAAGTGTCAACTCCAACTCTTGTAGAGGTCTCAAGAAACCTAGGAAAAGTGGGCAGCAAATGTTGTAAACATCCTGAAGCAAAAAGAATGCCCTGTGCAGAAGACTATCTATCCGTGGTCCTGAACCAGTTATGTGTGTTGCATGAGAAAACGCCAGTAAGTGACAGAGTCACCAAATGCTGCACAGAATCCTTGGTGAACAGGCGACCATGCTTTTCAGCTCTGGAAGTCGATGAAACATACGTTCCCAAAGAGTTTAATGCTGAAACGTTCACCTTCCATGCAGATATATGCACACTTTCTGAGAAGGAGAGACAAATCAAGAAACAAACTGCACTTGTTGAGCTTGTGAAACACAAGCCCAAGGCAACAAAAGAGCAACTGAAAGCTGTTATGGATGATTTCGCAGCTTTTGTAGAGAAGTGCTGCAAGGCTGACGATAAGGAGACCTGCTTTGCCGAGGAGGGTAAAAAACTTGTTGCTGCAAGTCAAGCTGCCTTAGGCTTATAACATCACATTTAAAAGCATCTCAGCCTACCATGAGAATAAGAGAAAGAAAATGAAGATCAAAAGCTTATTCATCTGTTTTTCTTTTTCGTTGGTGTAAAGCCAACACCCTGTCTAAAAAACATAAATTTCTTTAATCATTTTGCCTCTTTTCTCTGTGCTTCAATTAACAAAAAA start: 9 end: 2045
Align Title:gi|51476389|emb|CR749331.1| Homo sapiens mRNA; cDNA DKFZp779N1935 (from clone DKFZp779N1935)
Align Len: 2075
Score: 4027.0 Bits: 3632.37 E-val: 0.0
N.aligns:None Ident:2036 Pos.:2036 Gaps:0 Align len:2051
Strand: (None, None) Frame: (1, 1)
Query: TCTCTTCTGTCAACCCCACGCGCCTTTGGCACAATGAAGTGGGTAACCTTTATTTCCCTTCTTTTTCTCTTTAGCTCGGCTTATTCCAGGGGTGTGTTTCGTCGAGATGCACACAAGAGTGAGGTTGCTCATCGGTTTAAAGATTTGGGAGAAGAAAATTTCAAAGCCTTAGTGTTGATTGCCTTTGCTCAGTATCTTCAGCAGTGTCCATTTGAAGATCATGTAAAATTAGTGAATGAAGTAACTGAATTTGCAAAAACATGTGTTGCTGATGAGTCAGCTGAAAATTGTGACAAATCACTTCATACCCTTTTTGGAGACAAATTATGCACAGTTGCAACTCTTCGTGAAACCTATGGTGAAATGGCTGACTGCTGTGCAAAACAAGAACCTGAGAGAAATGAATGCTTCTTGCAACACAAAGATGACAACCCAAACCTCCCCCGATTGGTGAGACCAGAGGTTGATGTGATGTGCACTGCTTTTCATGACAATGAAGAGACATTTTTGAAAAAATACTTATATGAAATTGCCAGAAGACATCCTTACTTCTATGCCCCGGAACTCCTTTTCTTTGCTAAAAGGTATAAAGCTGCTTTTACAGAATGTTGCCAAGCTGCTGATAAGGCTGCCTGCCTGTTGCCAAAGCTCGATGAACTTCGGGATGAAGGGAAGGCTTCGTCTGCCAAACAGAGGCTCAAGTGTGCCAGTCTCCAAAAATTTGGAGAAAGAGCTTTCAAAGCATGGGCAGTAGCTCGCCTGAGCCAGAGATTTCCCAAAGCTGAGTTTGCAGAAGTTTCCAAGTTAGTGACAGATCTTACCAAAGTCCACACGGAATGCTGCCATGGAGATCTGCTTGAATGTGCTGATGACAGGGCGGACCTTGCCAAGTATATCTGTGAAAATCAAGATTCGATCTCCAGTAAACTGAAGGAATGCTGTGAAAAACCTCTGTTGGAAAAATCCCACTGCATTGCCGAAGTGGAAAATGATGAGATGCCTGCTGACTTGCCTTCATTAGCTGCTGATTTTGTTGAAAGTAAGGATGTTTGCAAAAACTATGCTGAGGCAAAGGATGTCTTCCTGGGCATGTTTTTGTATGAATATGCAAGAAGGCATCCTGATTACTCTGTCGTGCTGCTGCTGAGACTTGCCAAGACATATAAAACCACTCTAGAGAAGTGCTGTGCCGCTGCAGATCCTCATGAATGCTATGCCAAAGTGTTCGATGAATTTAAACCTCTTGTGGAAGAGCCTCAGAATTTAATCAAACAAAATTGTGAGCTTTTTGAGCAGCTTGGAGAGTACAAATTCCAGAATGCGCTATTAGTTCGTTACACCAAGAAAGTACCCCAAGTGTCAACTCCAACTCTTGTAGAGGTCTCAAGAAACCTAGGAAAAGTGGGCAGCAAATGTTGTAAACATCCTGAAGCAAAAAGAATGCCCTGTGCAGAAGACTATCTATCCGTGGTCCTGAACCAGTTATGTGTGTTGCATGAGAAAACGCCAGTAAGTGACAGAGTCACCAAATGCTGCACAGAATCCTTGGTGAACAGGCGACCATGCTTTTCAGCTCTGGAAGTCGATGAAACATACGTTCCCAAAGAGTTTAATGCTGAAACATTCACCTTCCATGCAGATATATGCACACTTTCTGAGAAGGAGAGACAAATCAAGAAACAAACTGCACTTGTTGAGCTTGTGAAACACAAGCCCAAGGCAACAAAAGAGCAACTGAAAGCTGTTATGGATGATTTCGCAGCTTTTGTAGAGAAGTGCTGCAAGGCTGACGATAAGGAGACCTGCTTTGCCGAGGAGGGTAAAAAACTTGTTGCTGCAAGTCGAGCTGCCTTAGGCTTATAACATCACATTTAAAAGCATCTCAGCCTACCATGAGAATAAGAGAAAGAAAATGAAGATCAAAAGCTTATTCATCTGTTTTTCTTTTTCGTTGGTGTAAAGCCAACACCCTGTCTAAAAAACATAAATTTCTTTAATCATTTTGCCTCTTTTCTCTGTGCTTCAATTAATAAAAAATGGAAAGAATCTAA start: 1 end: 2051
Match: ||||||||||||||||||| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||| ||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| ||||||||||||||||||||||||||| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Subjc: TCTCTTCTGTCAACCCCACACGCCTTTGGCACAATGAAGTGGGTAACCTTTATTTCCCTTCTTTTTCTCTTTAGCTCGGCTTATTCCAGGGGTGTGTTTCGTCGAGATGCATACAAGAGTGAGGTTGCTCATCGGTTTAAAGATTTGGGAGAAGAAAATTTCAAAGCCTTGGTGTTGATTGCCTTTGCTCAGTATCCTCAGCAGTGTCCATTTGAAGATCATGTAAAATTAGTGAATGAAGTAACTGAATTTGCAAAAACATGTGTTGCTGATGAGTCAGCTGAAAATTGTGACAAATCACTTCATACCCTTTTTGGAGACAAATTATGCACAGTTGCAACTCTTCGTGAAACCTATGGTGAAATGGCTGACTGCTGTGCAAAACAAGAACCTGAGAGAAATGAATGCTTCTTGCAACACAAAGATGACAACCCAAACCTCCCCCGATTGGTGAGACCAGAGGTTGATGTGATGTGCACTGCTTTTCATGACAATGAAGAGACATTTTTGAAAAAATACTTATATGAAATTGCCAGAAGACATCCTTACTTTTATGCCCCGGAACTCCTTTTCTTTGCTAAAAGGTATAAAGCTGCTTTTACAGAATGTTGCCAAGCTGCTGATAAAGCTGCCTGCCTGTTGCCAAAGCTCGATGAACTTCGGGATGAAGGGAAGGCTTCGTCTGCCAAACAGAGACTCAAGTGTGCCAGTCTCCAAAAATTTGGAGAAAGAGCTTTCAAAGCATGGGCAGTAGCTCGCCTGAGCCAGAGATTTCCCAAAGCTGAGTTTGCAGAAGTTTCCAAGTTAGTGACAGATCTTACCAAAGTCCACACGGAATGCTGCCATGGAGATCTGCTTGAATGTGCTGATGACAGGGCGGACCTTGCCAAGTATATCTGTGAAAATCAAGATTCGATCTCCAGTAAACTGAAGGAATGCTGCGAAAAACCTCTGTTGGAAAAATCCCACTGCATTGCCGAAGTGGAAAATGATGAGATGCCTGCTGACTTGCCTTCATTAGCTGCTGATTTTGTTGAAAGTAAGGATGTTTGCAAAAACTATGCTGAGGCAAAGGATGTCTTCCTGGGCATGTTTTTGTATGAATATGCAAGAAGGCATCCTGATTACTCTGTCGTGCTGCTGCTGAGACTTGCCAAGACATATGAAACCACTCTAGAGAAGTGCTGTGCCGCTGCAGATCCTCATGAATGCTATGCCAAAGTGTTCGATGAATTTAAACCTCTTGTGGAAGAGCCTCAGAATTTAATCAAACAAAATTGTGAGCTTTTTGAGCAGCTTGGAGAGTACAAATTCCAGAATGCGCTATTAGTTCGTTACACCAAGAAAGTACCCCAAGTGTCAACTCCAACTCTTGTAGAGGTCTCAAGAAACCTAGGAAAAGTGGGCAGCAAATGTTGTAAACATCCTGAAGCAAAAAGAATGCCCTGTGCAGAAGACTATCTATCCGTGGTCCTGAACCAGTTATGTGTGTTGCATGAGAGAACGCCAGTAAGTGACAGAGTCACCAAATGCTGCACAGAATCCTTGGTGAACAGGCGACCATGCTTTTCAGCTCTGGAAGTCGATGAAACATACGTTCCCAAAGAGTTTAATGCTGAAACATTCACCTTCCATGCAGATATATGCACACTTTCTGAGAAGGAGAGACAAATCAAGAAACAAGCTGCACTTGTTGAGCTTGTGAAACACAGGCCCAAGGCAACAAAAGAGCAACTGAAAGCTGTTATGGATGATTTCGCAGCTTTTGTAGAGAAGTGCTGCAAGGCTGACGATAAGGAGACCTGCTTTGCCGAGGAGGGTAAAAAACTTGTTGCTGCAAGTCAAGCTGCCTTAGGCTTATAACATTACATTTAAAAGCATCTCAGCCTACCATGAGAATAAGAGAAAGAAAATGAAGATCAAAAGCTTATTCATCTGTTTTTCTTTTTCGTTGGTGTAAAGCCAACACCCTGTCCAAAAAACATAAATTTCTTTAATCATTTTGCCTCTTTTCTCTGTGCTTCAATTAATAAAAAATGGAAAGAATCTAA start: 8 end: 2058
Align Title:gi|21706470|gb|BC034026.1| Homo sapiens cDNA clone IMAGE:4734794, containing frame-shift errors
Align Len: 2059
Score: 4010.0 Bits: 3617.04 E-val: 0.0
N.aligns:None Ident:2025 Pos.:2025 Gaps:1 Align len:2037
Strand: (None, None) Frame: (1, 1)
Query: TCTCTTCTGTCAACCCCACGCGCCTTTGGCACAATGAAGTGGGTAACCTTTATTTCCCTTCTTTTTCTCTTTAGCTCGGCTTATTCCAGGGGTGTGTTTCGTCGAGATGCACACAAGAGTGAGGTTGCTCATCGGTTTAAAGATTTGGGAGAAGAAAATTTCAAAGCCTTAGTGTTGATTGCCTTTGCTCAGTATCTTCAGCAGTGTCCATTTGAAGATCATGTAAAATTAGTGAATGAAGTAACTGAATTTGCAAAAACATGTGTTGCTGATGAGTCAGCTGAAAATTGTGACAAATCACTTCATACCCTTTTTGGAGACAAATTATGCACAGTTGCAACTCTTCGTGAAACCTATGGTGAAATGGCTGACTGCTGTGCAAAACAAGAACCTGAGAGAAATGAATGCTTCTTGCAACACAAAGATGACAACCCAAACCTCCCCCGATTGGTGAGACCAGAGGTTGATGTGATGTGCACTGCTTTTCATGACAATGAAGAGACATTTTTGAAAAAATACTTATATGAAATTGCCAGAAGACATCCTTACTTCTATGCCCCGGAACTCCTTTTCTTTGCTAAAAGGTATAAAGCTGCTTTTACAGAATGTTGCCAAGCTGCTGATAAGGCTGCCTGCCTGTTGCCAAAGCTCGATGAACTTCGGGATGAAGGGAAGGCTTCGTCTGCCAAACAGAGGCTCAAGTGTGCCAGTCTCCAAAAATTTGGAGAAAGAGCTTTCAAAGCATGGGCAGTAGCTCGCCTGAGCCAGAGATTTCCCAAAGCTGAGTTTGCAGAAGTTTCCAAGTTAGTGACAGATCTTACCAAAGTCCACACGGAATGCTGCCATGGAGATCTGCTTGAATGTGCTGATGACAGGGCGGACCTTGCCAAGTATATCTGTGAAAATCAAGATTCGATCTCCAGTAAACTGAAGGAATGCTGTGAAAAACCTCTGTTGGAAAAATCCCACTGCATTGCCGAAGTGGAAAATGATGAGATGCCTGCTGACTTGCCTTCATTAGCTGCTGATTTTGTTGAAAGTAAGGATGTTTGCAAAAACTATGCTGAGGCAAAGGATGTCTTCCTGGGCATGTTTTTGTATGAATATGCAAGAAGGCATCCTGATTACTCTGTCGTGCTGCTGCTGAGACTTGCCAAGACATATAAAACCACTCTAGAGAAGTGCTGTGCCGCTGCAGATCCTCATGAATGCTATGCCAAAGTGTTCGATGAATTTAAACCTCTTGTGGAAGAGCCTCAGAATTTAATCAAACAAAATTGTGAGCTTTTTGAGCAGCTTGGAGAGTACAAATTCCAGAATGCGCTATTAGTTCGTTACACCAAGAAAGTACCCCAAGTGTCAACTCCAACTCTTGTAGAGGTCTCAAGAAACCTAGGAAAAGTGGGCAGCAAATGTTGTAAACATCCTGAAGCAAAAAGAATGCCCTGTGCAGAAGACTATCTATCCGTGGTCCTGAACCAGTTATGTGTGTTGCATGAGAAAACGCCAGTAAGTGACAGAGTCACCAAATGCTGCACAGAATCCTTGGTGAACAGGCGACCATGCTTTTCAGCTCTGGAAGTCGATGAAACATACGTTCCCAAAGAGTTTAATGCTGAAACATTCACCTTCCATGCAGATATATGCACACTTTCTGAGAAGGAGAGACAAATCAAGAAACAAACTGCACTTGTTGAGCTTGTGAAACACAAGCCCAAGGCAACAAAAGAGCAACTGAAAGCTGTTATGGATGATTTCGCAGCTTTTGTAGAGAAGTGCTGCAAGGCTGACGATAAGGAGACCTGCTTTGCCGAGGAGGGTAAAAAACTTGTTGCTGCAAGTCGAGCTGCCTTAGGCTTATAACATCACATTTAAAAGCATCTCAGCCTACCATGAGAATAAGAGAAAGAAAATGAAGATCAAAAGCTTATTCATCTGTTTTTCTTTTTCGTTGGTGTAAAGCCAACACCCTGTCTAAAAAACATAAATTTCTTTAATCATTTTGCCTCTTTTCTCTGTGCTTCAATTAATAAAAAA start: 1 end: 2037
Match: ||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| || ||||||
Subjc: TCTCTTCTGTCAACCCCACACGCCTTTGGCACAATGAAGTGGGTAACCTTTATTTCCCTTCTTTTTCTCTTTAGCTCGGCTTATTCCAGGGGTGTGTTTCGTCGAGATGCACACAAGAGTGAGGTTGCTCATCGGTTTAAAGATTTGGGAGAAGAAAATTTCAAAGCCTTGGTGTTGATTGCCTTTGCTCAGTATCTTCAGCAGTGTCCATTTGAAGATCATGTAAAATTAGTGAATGAAGTAACTGAATTTGCAAAAACATGTGTTGCTGATGAGTCAGCTGAAAATTGTGACAAATCACTTCATACCCTTTTTGGAGACAA-TTATGCACAGTTGCAACTCTTCGTGAAACCTATGGTGAAATGGCTGACTGCTGTGCAAAACAAGAACCTGAGAGAAATGAATGCTTCTTGCAACACAAAGATGACAACCCAAACCTCCCCCGATTGGTGAGACCAGAGGTTGATGTGATGTGCACTGCTTTTCATGACAATGAAGAGACATTTTTGAAAAAATACTTATATGAAATTGCCAGAAGACATCCTTACTTTTATGCCCCGGAACTCCTTTTCTTTGCTAAAAGGTATAAAGCTGCTTTTACAGAATGTTGCCAAGCTGCTGATAAAGCTGCCTGCCTGTTGCCAAAGCTCGATGAACTTCGGGATGAAGGGAAGGCTTCGTCTGCCAAACAGAGACTCAAGTGTGCCAGTCTCCAAAAATTTGGAGAAAGAGCTTTCAAAGCATGGGCAGTAGCTCGCCTGAGCCAGAGATTTCCCAAAGCTGAGTTTGCAGAAGTTTCCAAGTTAGTGACAGATCTTACCAAAGTCCACACGGAATGCTGCCATGGAGATCTGCTTGAATGTGCTGATGACAGGGCGGACCTTGCCAAGTATATCTGTGAAAATCAAGATTCGATCTCCAGTAAACTGAAGGAATGCTGTGAAAAACCTCTGTTGGAAAAATCCCACTGCATTGCCGAAGTGGAAAATGATGAGATGCCTGCTGACTTGCCTTCATTAGCTGCTGATTTTGTTGAAAGTAAGGATGTTTGCAAAAACTATGCTGAGGCAAAGGATGTCTTCCTGGGCATGTTTTTGTATGAATATGCAAGAAGGCATCCTGATTACTCTGTCGTGCTGCTGCTGAGACTTGCCAAGACATATGAAACCACTCTAGAGAAGTGCTGTGCCGCTGCAGATCCTCATGAATGCTATGCCAAAGTGTTCGATGAATTTAAACCTCTTGTGGAAGAGCCTCAGAATTTAATCAAACAAAATTGTGAGCTTTTTGAGCAGCTTGGAGAGTACAAATTCCAGAATGCGCTATTAGTTCGTTACACCAAGAAAGTACCCCAAGTGTCAACTCCAACTCTTGTAGAGGTCTCAAGAAACCTAGGAAAAGTGGGCAGCAAATGTTGTAAACATCTTGAAGCAAAAAGAATGCCCTGTGCAGAAGACTATCTATCCGTGGTCCTGAACCAGTTATGTGTGTTGCATGAGAAAACGCCAGTAAGTGACAGAGTCACCAAATGCTGCACAGAATCCTTGGTGAACAGGCGACCATGCTTTTCAGCTCTGGAAGTCGATGAAACATACGTTCCCAAAGAGTTTAATGCTGAAACATTCACCTTCCATGCAGATATATGCACACTTTCTGAGAAGGAGAGACAAATCAAGAAACAAACTGCACTTGTTGAGCTTGTGAAACACAAGCCCAAGGCAACAAAAGAGCAACTGAAAGCTGTTATGGATGATTTCGCAGCTTTTGTAGAGAAGTGCTGCAAGGCTGACGATAAGGAGACCTGCTTTGCCGAGGAGGGTAAAAAACTTGTTGCTGCAAGTCAAGCTGCCTTAGGCTTATAACATCACATTTAAAAGCATCTCAGCCTACCATGAGAATAAGAGAAAGAAAATGAAGATCAAAAGCTTATTCATCTGTTTTTCTTTTTCGTTGGTGTAAAGCCAACACCCTGTCTAAAAAACATAAATTTCTTTAATCATTTTGCCTCTTTTCTCTGTGCTTCAAAAAAAAAAAAA start: 7 end: 2042
Align Title:gi|194391079|dbj|AK298437.1| Homo sapiens cDNA FLJ54371 complete cds, highly similar to Serum albumin precursor
Align Len: 2082
Score: 3994.0 Bits: 3602.61 E-val: 0.0
N.aligns:None Ident:2040 Pos.:2040 Gaps:27 Align len:2076
Strand: (None, None) Frame: (1, 1)
Query: TCTCTTCTGTCAACCCCACGCGCCTTTGGCACAATGAAGTGGGTAACCTTTATTTCCCTTCTTTTTCTCTTTAGCTCGGCTTATTCCAGGGGTGTGTTTCGTCGAGATGCAC---------------------------ACAAGAGTGAGGTTGCTCATCGGTTTAAAGATTTGGGAGAAGAAAATTTCAAAGCCTTAGTGTTGATTGCCTTTGCTCAGTATCTTCAGCAGTGTCCATTTGAAGATCATGTAAAATTAGTGAATGAAGTAACTGAATTTGCAAAAACATGTGTTGCTGATGAGTCAGCTGAAAATTGTGACAAATCACTTCATACCCTTTTTGGAGACAAATTATGCACAGTTGCAACTCTTCGTGAAACCTATGGTGAAATGGCTGACTGCTGTGCAAAACAAGAACCTGAGAGAAATGAATGCTTCTTGCAACACAAAGATGACAACCCAAACCTCCCCCGATTGGTGAGACCAGAGGTTGATGTGATGTGCACTGCTTTTCATGACAATGAAGAGACATTTTTGAAAAAATACTTATATGAAATTGCCAGAAGACATCCTTACTTCTATGCCCCGGAACTCCTTTTCTTTGCTAAAAGGTATAAAGCTGCTTTTACAGAATGTTGCCAAGCTGCTGATAAGGCTGCCTGCCTGTTGCCAAAGCTCGATGAACTTCGGGATGAAGGGAAGGCTTCGTCTGCCAAACAGAGGCTCAAGTGTGCCAGTCTCCAAAAATTTGGAGAAAGAGCTTTCAAAGCATGGGCAGTAGCTCGCCTGAGCCAGAGATTTCCCAAAGCTGAGTTTGCAGAAGTTTCCAAGTTAGTGACAGATCTTACCAAAGTCCACACGGAATGCTGCCATGGAGATCTGCTTGAATGTGCTGATGACAGGGCGGACCTTGCCAAGTATATCTGTGAAAATCAAGATTCGATCTCCAGTAAACTGAAGGAATGCTGTGAAAAACCTCTGTTGGAAAAATCCCACTGCATTGCCGAAGTGGAAAATGATGAGATGCCTGCTGACTTGCCTTCATTAGCTGCTGATTTTGTTGAAAGTAAGGATGTTTGCAAAAACTATGCTGAGGCAAAGGATGTCTTCCTGGGCATGTTTTTGTATGAATATGCAAGAAGGCATCCTGATTACTCTGTCGTGCTGCTGCTGAGACTTGCCAAGACATATAAAACCACTCTAGAGAAGTGCTGTGCCGCTGCAGATCCTCATGAATGCTATGCCAAAGTGTTCGATGAATTTAAACCTCTTGTGGAAGAGCCTCAGAATTTAATCAAACAAAATTGTGAGCTTTTTGAGCAGCTTGGAGAGTACAAATTCCAGAATGCGCTATTAGTTCGTTACACCAAGAAAGTACCCCAAGTGTCAACTCCAACTCTTGTAGAGGTCTCAAGAAACCTAGGAAAAGTGGGCAGCAAATGTTGTAAACATCCTGAAGCAAAAAGAATGCCCTGTGCAGAAGACTATCTATCCGTGGTCCTGAACCAGTTATGTGTGTTGCATGAGAAAACGCCAGTAAGTGACAGAGTCACCAAATGCTGCACAGAATCCTTGGTGAACAGGCGACCATGCTTTTCAGCTCTGGAAGTCGATGAAACATACGTTCCCAAAGAGTTTAATGCTGAAACATTCACCTTCCATGCAGATATATGCACACTTTCTGAGAAGGAGAGACAAATCAAGAAACAAACTGCACTTGTTGAGCTTGTGAAACACAAGCCCAAGGCAACAAAAGAGCAACTGAAAGCTGTTATGGATGATTTCGCAGCTTTTGTAGAGAAGTGCTGCAAGGCTGACGATAAGGAGACCTGCTTTGCCGAGGAGGGTAAAAAACTTGTTGCTGCAAGTCGAGCTGCCTTAGGCTTATAACATCACATTTAAAAGCATCTCAGCCTACCATGAGAATAAGAGAAAGAAAATGAAGATCAAAAGCTTATTCATCTGTTTTTCTTTTTCGTTGGTGTAAAGCCAACACCCTGTCTAAAAAACATAAATTTCTTTAATCATTTTGCCTCTTTTCTCTGTGCTTCAATTAATAAAAAATGGAAAGAATCT start: 1 end: 2049
Match: ||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Subjc: TCTCTTCTGTCAACCCCACACGCCTTTGGCACAATGAAGTGGGTAACCTTTATTTCCCTTCTTTTTCTCTTTAGCTCGGCTTATTCCAGGGGTGTGTTTCGTCGAGATGCACTATTTATTTTTTTCTTCCCTTGCCCAGACAAGAGTGAGGTTGCTCATCGGTTTAAAGATTTGGGAGAAGAAAATTTCAAAGCCTTGGTGTTGATTGCCTTTGCTCAGTATCTTCAGCAGTGTCCATTTGAAGATCATGTAAAATTAGTGAATGAAGTAACTGAATTTGCAAAAACATGTGTTGCTGATGAGTCAGCTGAAAATTGTGACAAATCACTTCATACCCTTTTTGGAGACAAATTATGCACAGTTGCAACTCTTCGTGAAACCTATGGTGAAATGGCTGACTGCTGTGCAAAACAAGAACCTGAGAGAAATGAATGCTTCTTGCAACACAAAGATGACAACCCAAACCTCCCCCGATTGGTGAGACCAGAGGTTGATGTGATGTGCACTGCTTTTCATGACAATGAAGAGACATTTTTGAAAAAATACTTATATGAAATTGCCAGAAGACATCCTTACTTTTATGCCCCGGAACTCCTTTTCTTTGCTAAAAGGTATAAAGCTGCTTTTACAGAATGTTGCCAAGCTGCTGATAAAGCTGCCTGCCTGTTGCCAAAGCTCGATGAACTTCGGGATGAAGGGAAGGCTTCGTCTGCCAAACAGAGACTCAAGTGTGCCAGTCTCCAAAAATTTGGAGAAAGAGCTTTCAAAGCATGGGCAGTAGCTCGCCTGAGCCAGAGATTTCCCAAAGCTGAGTTTGCAGGAGTTTCCAAGTTAGTGACAGATCTTACCAAAGTCCACACGGAATGCTGCCATGGAGATCTGCTTGAATGTGCTGATGACAGGGCGGACCTTGCCAAGTATATCTGTGAAAATCAAGATTCGATCTCCAGTAAACTGAAGGAATGCTGTGAAAAACCTCTGTTGGAAAAATCCCACTGCATTGCCGAAGTGGAAAATGATGAGATGCCTGCTGACTTGCCTTCATTAGCTGCTGATTTTGTTGAAAGTAAGGATGTTTGCAAAAACTATGCTGAGGCAAAGGATGTCTTCCTGGGCATGTTTTTGTATGAATATGCAAGAAGGCATCCTGATTACTCTGTCGTGCTGCTGCTGAGACTTGCCAAGACATATGAAACCACTCTAGAGAAGTGCTGTGCCGCTGCAGATCCTCATGAATGCTATGCCAAAGTGTTCGATGAATTTAAACCTCTTGTGGAAGAGCCTCAGAATTTAATCAAACAAAATTGTGAGCTTTTTGAGCAGCTTGGAGAGTACAAATTCCAGAATGCGCTATTAGTTCGTTACACCAAGAAAGTACCCCAAGTGTCAACTCCAACTCTTGTAGAGGTCTCAAGAAACCTAGGAAAAGTGGGCAGCAAATGTTGTAAACATCCTGAAGCAAAAAGAATGCCCTGTGCAGAAGACTATCTATCCGTGGTCCTGAACCAGTTATGTGTGTTGGATGAGAAAACGCCAGTAAGTGACAGAGTCACCAAATGCTGCACAGAATCCTTGGTGAACAGGCGACCATGCTTTTCAGCTCTGGAAGTCGATGAAACATACGTTCCCAAAGAGTTTAATGCTGAAACATTCACCTTCCATGCAGATATATGCACACTTTCTGAGAAGGAGAGACAAATCAAGAAACAAACTGCACTTGTTGAGCTTGTGAAACACAAGCCCAAGGCAACAAAAGAGCAACTGAAAGCTGTTATGGATGATTTCGCAGCTTTTGTAGAGAAGTGCTGCAAGGCTGACGATAAGGAGACCTGCTTTGCCGAGGAGGGTAAAAAACTTGTTGCTGCAAGTCAAGCTGCCTTAGGCTTATAACATCACATTTAAAAGCATCTCAGCCTACCATGAGAATAAGAGAAAGAAAATGAAGATCAAAAGCTTATTCATCTGTTTTTCTTTTTCGTTGGTGTAAAGCCAACACCCTGTCTAAAAAACATAAATTTCTTTAATCATTTTGCCTCTTTTCTCTGTGCTTCAATTAATAAAAAATGGAAAGAATCT start: 7 end: 2082
Align Title:gi|408407943|gb|AF130077.2| Homo sapiens clone FLB9714 PRO2619 mRNA, complete cds
Align Len: 2560
Score: 3937.0 Bits: 3551.22 E-val: 0.0
N.aligns:None Ident:1982 Pos.:1982 Gaps:0 Align len:1991
Strand: (None, None) Frame: (1, 1)
Query: CTTTTTCTCTTTAGCTCGGCTTATTCCAGGGGTGTGTTTCGTCGAGATGCACACAAGAGTGAGGTTGCTCATCGGTTTAAAGATTTGGGAGAAGAAAATTTCAAAGCCTTAGTGTTGATTGCCTTTGCTCAGTATCTTCAGCAGTGTCCATTTGAAGATCATGTAAAATTAGTGAATGAAGTAACTGAATTTGCAAAAACATGTGTTGCTGATGAGTCAGCTGAAAATTGTGACAAATCACTTCATACCCTTTTTGGAGACAAATTATGCACAGTTGCAACTCTTCGTGAAACCTATGGTGAAATGGCTGACTGCTGTGCAAAACAAGAACCTGAGAGAAATGAATGCTTCTTGCAACACAAAGATGACAACCCAAACCTCCCCCGATTGGTGAGACCAGAGGTTGATGTGATGTGCACTGCTTTTCATGACAATGAAGAGACATTTTTGAAAAAATACTTATATGAAATTGCCAGAAGACATCCTTACTTCTATGCCCCGGAACTCCTTTTCTTTGCTAAAAGGTATAAAGCTGCTTTTACAGAATGTTGCCAAGCTGCTGATAAGGCTGCCTGCCTGTTGCCAAAGCTCGATGAACTTCGGGATGAAGGGAAGGCTTCGTCTGCCAAACAGAGGCTCAAGTGTGCCAGTCTCCAAAAATTTGGAGAAAGAGCTTTCAAAGCATGGGCAGTAGCTCGCCTGAGCCAGAGATTTCCCAAAGCTGAGTTTGCAGAAGTTTCCAAGTTAGTGACAGATCTTACCAAAGTCCACACGGAATGCTGCCATGGAGATCTGCTTGAATGTGCTGATGACAGGGCGGACCTTGCCAAGTATATCTGTGAAAATCAAGATTCGATCTCCAGTAAACTGAAGGAATGCTGTGAAAAACCTCTGTTGGAAAAATCCCACTGCATTGCCGAAGTGGAAAATGATGAGATGCCTGCTGACTTGCCTTCATTAGCTGCTGATTTTGTTGAAAGTAAGGATGTTTGCAAAAACTATGCTGAGGCAAAGGATGTCTTCCTGGGCATGTTTTTGTATGAATATGCAAGAAGGCATCCTGATTACTCTGTCGTGCTGCTGCTGAGACTTGCCAAGACATATAAAACCACTCTAGAGAAGTGCTGTGCCGCTGCAGATCCTCATGAATGCTATGCCAAAGTGTTCGATGAATTTAAACCTCTTGTGGAAGAGCCTCAGAATTTAATCAAACAAAATTGTGAGCTTTTTGAGCAGCTTGGAGAGTACAAATTCCAGAATGCGCTATTAGTTCGTTACACCAAGAAAGTACCCCAAGTGTCAACTCCAACTCTTGTAGAGGTCTCAAGAAACCTAGGAAAAGTGGGCAGCAAATGTTGTAAACATCCTGAAGCAAAAAGAATGCCCTGTGCAGAAGACTATCTATCCGTGGTCCTGAACCAGTTATGTGTGTTGCATGAGAAAACGCCAGTAAGTGACAGAGTCACCAAATGCTGCACAGAATCCTTGGTGAACAGGCGACCATGCTTTTCAGCTCTGGAAGTCGATGAAACATACGTTCCCAAAGAGTTTAATGCTGAAACATTCACCTTCCATGCAGATATATGCACACTTTCTGAGAAGGAGAGACAAATCAAGAAACAAACTGCACTTGTTGAGCTTGTGAAACACAAGCCCAAGGCAACAAAAGAGCAACTGAAAGCTGTTATGGATGATTTCGCAGCTTTTGTAGAGAAGTGCTGCAAGGCTGACGATAAGGAGACCTGCTTTGCCGAGGAGGGTAAAAAACTTGTTGCTGCAAGTCGAGCTGCCTTAGGCTTATAACATCACATTTAAAAGCATCTCAGCCTACCATGAGAATAAGAGAAAGAAAATGAAGATCAAAAGCTTATTCATCTGTTTTTCTTTTTCGTTGGTGTAAAGCCAACACCCTGTCTAAAAAACATAAATTTCTTTAATCATTTTGCCTCTTTTCTCTGTGCTTCAATTAATAAAAAATGGAAAGAATCTAA start: 61 end: 2051
Match: || || |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| ||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Subjc: CTGTTACTCTTTAGCTCGGCTTATTCCAGGGGTGTGTTTCGTCGAGATGCACACAAGAGTGAGGTTGCTCATCGGTTTAAAGATTTGGGAGAAGAAAATTTCAAAGCCTTGGTGTTGATTGCCTTTGCTCAGTATCTTCAGCAGTGTCCATTTGAAGATCATGTAAAATTAGTGAATGAAGTAACTGAATTTGCAAAAACATGTGTTGCTGATGAGTCAGCTGAAAATTGTGACAAATCACTTCATACCCTTTTTGGAGACAAATTATGCACAGTTGCAACTCTTCGTGAAACCTATGGTGAAATGGCTGACTGCTGTGCAAAACAAGAACCTGAGAGAAATGAATGCTTCTTGCAACACAAAGATGACAACCCAAACCTCCCCCGATTGGTGAGACCAGAGGTTGATGTGATGTGCACTGCTTTTCATGACAATGAAGAGACATTTTTGAAAAAATACTTATATGAAATTGCCAGAAGACATCCTTACTTTTATGCCCCGGAACTCCTTTTCTTTGCTAAAAGGTATAAAGCTGCTTTTACAGAATGTTGCCAAGCTGCTGATAAAGCTGCCTGCCTGTTGCCAAAGCTCGATGAACTTCGGGATGAAGGGAAGGCTTCGTCTGCCAAACAGAGACTCAAGTGTGCCAGTCTCCAAAAATTTGGAGAAAGAGCTTTCAAAGCATGGGCAGTAGCTCGCCTGAGCCAGAGATTTCCCAAAGCTGAGTTTGCAGAAGTTTCCAAGTTAGTGACAGATCTTACCAAAGTCCACACGGAATGCTGCCATGGAGATCTGCTTGAATGTGCTGATGACAGGGCGGACCTTGCCAAGTATATCTGTGAAAATCAAGATTCGATCTCCAGTAAACTGAAGGAATGCTGTGAAAAACCTCTGTTGGAAAAATCCCACTGCATTGCCGAAGTGGAAAATGATGAGATGCCTGCTGACTTGCCTTCATTAGCTGCTGATTTTGTTGAAAGTAAGGATGTTTGCAAAAACTATGCTGAGGCAAAGGATGTCTTCCTGGGCATGTTTTTGTATGAATATGCAAGAAGGCATCCTGATTACTCTGTCGTGCTGCTGCTGAGACTTGCCAAGACATATGAAACCACTCTAGAGAAGTGCTGTGCCGCTGCAGATCCTCATGAATGCTATGCCAAAGTGTTCGATGAATTTAAACCTCTTGTGGAAGAGCCTCAGAATTTAATCAAACAAAATTGTGAGCTTTTTGAGCAGCTTGGAGAGTACAAATTCCAGAATGCGCTATTAGTTCGTTACACCAAGAAAGTACCCCAAGTGTCAACTCCAACTCTTGTAGAGGTCTCAAGAAACCTAGGAAAAGTGGGCAGCAAATGTTGTAAACATCCTGAAGCAAAAAGAATGCCCTGTGCAGAAGACTATCTATCCGTGGTCCTGAACCAGTTATGTGTGTTGCATGAGAAAACGCCAGTAAGTGACAGAGTCACCAAATGCTGCACAGAATCCTTGGTGAACAGGCGACCATGCTTTTCAGCTCTGGAAGTCGATGAAACATACGTTCCCAAAGAGTTTAATGCTGAAACATTCACCTTCCATGCAGATATATGCACACTTTCTGAGAAGGAGAGACAAATCAAGAAACAAACTGCACTTGTTGAGCTTGTGAAACACAAGCCCAAGGCAACAAAAGAGCAACTGAAAGCTGTTATGGATGATTTCGCAGCTTTTGTAGAGAAGTGCTGCAAGGCTGACGATAAGGAGACCTGCTTTGCCGAGGAGGGTAAAAAACTTGTTGCTGCAAGTCAAGCTGCCTTAGGCTTATAACATCACATTTAAAAGCATCTCAGCCTACCACGAGAATAAGAGAAAGAAAATGAAGATCAAAAGCTTATTCATCTGTTTTTCTTTTTCGTTGGTGTAAAGCCAACACCCTGTCTAAAAAACATAAATTTCTTTAATCATTTTGCCTCTTTTCTCTGTGCTTCAATTAATAAAAAATGGAAAGAATCTAA start: 477 end: 2467
Align Title:gi|62113340|gb|AY960291.1| Homo sapiens serum albumin mRNA, complete cds
Align Len: 1981
Score: 3907.0 Bits: 3524.17 E-val: 0.0
N.aligns:None Ident:1970 Pos.:1970 Gaps:0 Align len:1981
Strand: (None, None) Frame: (1, 1)
Query: CTCTTCTGTCAACCCCACGCGCCTTTGGCACAATGAAGTGGGTAACCTTTATTTCCCTTCTTTTTCTCTTTAGCTCGGCTTATTCCAGGGGTGTGTTTCGTCGAGATGCACACAAGAGTGAGGTTGCTCATCGGTTTAAAGATTTGGGAGAAGAAAATTTCAAAGCCTTAGTGTTGATTGCCTTTGCTCAGTATCTTCAGCAGTGTCCATTTGAAGATCATGTAAAATTAGTGAATGAAGTAACTGAATTTGCAAAAACATGTGTTGCTGATGAGTCAGCTGAAAATTGTGACAAATCACTTCATACCCTTTTTGGAGACAAATTATGCACAGTTGCAACTCTTCGTGAAACCTATGGTGAAATGGCTGACTGCTGTGCAAAACAAGAACCTGAGAGAAATGAATGCTTCTTGCAACACAAAGATGACAACCCAAACCTCCCCCGATTGGTGAGACCAGAGGTTGATGTGATGTGCACTGCTTTTCATGACAATGAAGAGACATTTTTGAAAAAATACTTATATGAAATTGCCAGAAGACATCCTTACTTCTATGCCCCGGAACTCCTTTTCTTTGCTAAAAGGTATAAAGCTGCTTTTACAGAATGTTGCCAAGCTGCTGATAAGGCTGCCTGCCTGTTGCCAAAGCTCGATGAACTTCGGGATGAAGGGAAGGCTTCGTCTGCCAAACAGAGGCTCAAGTGTGCCAGTCTCCAAAAATTTGGAGAAAGAGCTTTCAAAGCATGGGCAGTAGCTCGCCTGAGCCAGAGATTTCCCAAAGCTGAGTTTGCAGAAGTTTCCAAGTTAGTGACAGATCTTACCAAAGTCCACACGGAATGCTGCCATGGAGATCTGCTTGAATGTGCTGATGACAGGGCGGACCTTGCCAAGTATATCTGTGAAAATCAAGATTCGATCTCCAGTAAACTGAAGGAATGCTGTGAAAAACCTCTGTTGGAAAAATCCCACTGCATTGCCGAAGTGGAAAATGATGAGATGCCTGCTGACTTGCCTTCATTAGCTGCTGATTTTGTTGAAAGTAAGGATGTTTGCAAAAACTATGCTGAGGCAAAGGATGTCTTCCTGGGCATGTTTTTGTATGAATATGCAAGAAGGCATCCTGATTACTCTGTCGTGCTGCTGCTGAGACTTGCCAAGACATATAAAACCACTCTAGAGAAGTGCTGTGCCGCTGCAGATCCTCATGAATGCTATGCCAAAGTGTTCGATGAATTTAAACCTCTTGTGGAAGAGCCTCAGAATTTAATCAAACAAAATTGTGAGCTTTTTGAGCAGCTTGGAGAGTACAAATTCCAGAATGCGCTATTAGTTCGTTACACCAAGAAAGTACCCCAAGTGTCAACTCCAACTCTTGTAGAGGTCTCAAGAAACCTAGGAAAAGTGGGCAGCAAATGTTGTAAACATCCTGAAGCAAAAAGAATGCCCTGTGCAGAAGACTATCTATCCGTGGTCCTGAACCAGTTATGTGTGTTGCATGAGAAAACGCCAGTAAGTGACAGAGTCACCAAATGCTGCACAGAATCCTTGGTGAACAGGCGACCATGCTTTTCAGCTCTGGAAGTCGATGAAACATACGTTCCCAAAGAGTTTAATGCTGAAACATTCACCTTCCATGCAGATATATGCACACTTTCTGAGAAGGAGAGACAAATCAAGAAACAAACTGCACTTGTTGAGCTTGTGAAACACAAGCCCAAGGCAACAAAAGAGCAACTGAAAGCTGTTATGGATGATTTCGCAGCTTTTGTAGAGAAGTGCTGCAAGGCTGACGATAAGGAGACCTGCTTTGCCGAGGAGGGTAAAAAACTTGTTGCTGCAAGTCGAGCTGCCTTAGGCTTATAACATCACATTTAAAAGCATCTCAGCCTACCATGAGAATAAGAGAAAGAAAATGAAGATCAAAAGCTTATTCATCTGTTTTTCTTTTTCGTTGGTGTAAAGCCAACACCCTGTCTAAAAAA start: 2 end: 1982
Match: ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Subjc: CTCTTCTGTCAACCCCACGCGCCTTTGGCACAATGAAGTGGGTAACCTTTATTTCCCTTCTTTTTCTCTTTAGCTCGGCTTATTCCAGGGGTGTGTTTCGTCGAGATGCACACAAGAGTGAGGTTGCTCATCGGTTTAAAGATTTGGGAGAAGAAAATTTCAAAGCCTTGGTGTTGATTGCCTTTGCTCAGTATCTTCAGCAGTGTCCATTTGAAGATCATGTAAAATTAGTGAATGAAGTAACTGAATTTGCAAAAACATGTGTTGCTGATGAGTCAGCTGAAAATTGTGACAAATCACTTCATACCCTTTTTGGAGACAAATTATGCACAGTTGCAACTCTTCGTGAAACCTATGGTGAAATGGCTGACTGCTGTGCAAAACAAGAACCTGAGAGAAATGAATGCTTCTTGCAACACAAAGATGACAACCCAAACCTCCCCCGATTGGTGAGACCAGAGGTTGATGTGATGTGCACTGCTTTTCATGACAATGAAGAGACATTTTTGAAAAAATACTTATATGAAATTGCCAGAAGACATCCTTACTTTTATGCCCCGGAACTCCTTTTCTTTGCTAAAAGGTATAAAGCTGCTTTTACAGAATGTTGCCAAGCTGCTGATAAAGCTGCCTGCCTGTTGCCAAAGCTCGATGAACTTCGGGATGAAGGGAAGGCTTCGTCTGCCAAACAGGGACTCAAGTGTGCCAGTCTCCAAAAATTTGGAGAAAGAGCTTTCAAAGCATGGGCAGTAGCTCGCCTGAGCCAGAGATTTCCCAAAGCTGAGTTTGCAGAAGTTTCCAAGTTAGTGACAGATCTTACCAAAGTCCACACGGAATGCTGCCATGGAGATCTGCTTGAATGTGCTGATGACAGGGCGGACCTTGCCAAGTATATCTGTGAAAATCAAGATTCGATCTCCAGTAAACTGAAGGAATGCTGTGAAAAACCTCTGTTGGAAAAATCCCACTGCATTGCCGAAGTGGAAAATGATGAGATGCCTGCTGACTTGCCTTCATTAGCTGCTGATTTTGTTGGAAGTAAGGATGTTTGCAAAAACTATGCTGAGGCAAAGGATGTCTTCCTGGGCATGTTTTTGTATGAATATGCAAGAAGGCATCCTGATTACTCTGTCGTGCTGCTGCTGAGACTTGCCAAGACATATGAAACCACTCTAGAGAAGTGCTGTGCCGCTGCAGATCCTCATGAATGCTATGCCAAAGTGTTCGATGAATTTAAACCTCTTGTGGAAGAGCCTCAGAATTTAATCAAACAAAATTGTGAGCTTTTTGAGCAGCTTGGAGAGTACAAATTCCAGAATGCGCTATTAGTTCGTTACACCAAGAAAGTACCCCAAGTGTCAACTCCAACTCTTGTAGAGGTCTCAAGAAACCTAGGAAAAGTGGGCAGCAAATGTTGTAAACATCCTGAAGCAAAAAGAATGCCCTGTGCAGAAGACTGTCTATCCGTGTTCCTGAACCAGTTATGTGTGTTGCATGAGAAAACGCCAGTAAGTGACAGAGTCACCAAATGCTGCACAGAATCCTTGGTGAACGGGCGACCATGCTTTTCAGCTCTGGAAGTCGATGAAACATACGTTCCCAAAGAGTTTAATGCTGAAACATTCACCTTCCATGCAGATATATGCACACTTTCTGAGAAGGAGAGACAAATCAAGAAACAAACTGCACTTGTTGAGCTTGTGAAACACAAGCCCAAGGCAACAAAAGAGCAACTGAAAGCTGTTATGGATGATTTCGCAGCTTTTGTAGAGAAGTGCTGCAAGGCTGACGATAAGGAGACCTGCTTTGCCGAGGAGGGTAAAAAACTTGTTGCTGCAAGTCAAGCTGCCTTAGGCTTATAACATCACATTTAAAAGCATCTCAGCCTACCATGAGAATAAGAGAAAGAAAATGAAGATCAAAAGCTTATTCATCTGTTTTTCTTTTTCGTTGGTGTAAAGCCAACACCCTGTCTAAAAAA start: 1 end: 1981
Align Title:gi|49176516|gb|AY550967.1| Homo sapiens cell growth inhibiting protein 42 mRNA, complete cds
Align Len: 1939
Score: 3848.0 Bits: 3470.97 E-val: 0.0
N.aligns:None Ident:1933 Pos.:1933 Gaps:0 Align len:1939
Strand: (None, None) Frame: (1, 1)
Query: TGGCACAATGAAGTGGGTAACCTTTATTTCCCTTCTTTTTCTCTTTAGCTCGGCTTATTCCAGGGGTGTGTTTCGTCGAGATGCACACAAGAGTGAGGTTGCTCATCGGTTTAAAGATTTGGGAGAAGAAAATTTCAAAGCCTTAGTGTTGATTGCCTTTGCTCAGTATCTTCAGCAGTGTCCATTTGAAGATCATGTAAAATTAGTGAATGAAGTAACTGAATTTGCAAAAACATGTGTTGCTGATGAGTCAGCTGAAAATTGTGACAAATCACTTCATACCCTTTTTGGAGACAAATTATGCACAGTTGCAACTCTTCGTGAAACCTATGGTGAAATGGCTGACTGCTGTGCAAAACAAGAACCTGAGAGAAATGAATGCTTCTTGCAACACAAAGATGACAACCCAAACCTCCCCCGATTGGTGAGACCAGAGGTTGATGTGATGTGCACTGCTTTTCATGACAATGAAGAGACATTTTTGAAAAAATACTTATATGAAATTGCCAGAAGACATCCTTACTTCTATGCCCCGGAACTCCTTTTCTTTGCTAAAAGGTATAAAGCTGCTTTTACAGAATGTTGCCAAGCTGCTGATAAGGCTGCCTGCCTGTTGCCAAAGCTCGATGAACTTCGGGATGAAGGGAAGGCTTCGTCTGCCAAACAGAGGCTCAAGTGTGCCAGTCTCCAAAAATTTGGAGAAAGAGCTTTCAAAGCATGGGCAGTAGCTCGCCTGAGCCAGAGATTTCCCAAAGCTGAGTTTGCAGAAGTTTCCAAGTTAGTGACAGATCTTACCAAAGTCCACACGGAATGCTGCCATGGAGATCTGCTTGAATGTGCTGATGACAGGGCGGACCTTGCCAAGTATATCTGTGAAAATCAAGATTCGATCTCCAGTAAACTGAAGGAATGCTGTGAAAAACCTCTGTTGGAAAAATCCCACTGCATTGCCGAAGTGGAAAATGATGAGATGCCTGCTGACTTGCCTTCATTAGCTGCTGATTTTGTTGAAAGTAAGGATGTTTGCAAAAACTATGCTGAGGCAAAGGATGTCTTCCTGGGCATGTTTTTGTATGAATATGCAAGAAGGCATCCTGATTACTCTGTCGTGCTGCTGCTGAGACTTGCCAAGACATATAAAACCACTCTAGAGAAGTGCTGTGCCGCTGCAGATCCTCATGAATGCTATGCCAAAGTGTTCGATGAATTTAAACCTCTTGTGGAAGAGCCTCAGAATTTAATCAAACAAAATTGTGAGCTTTTTGAGCAGCTTGGAGAGTACAAATTCCAGAATGCGCTATTAGTTCGTTACACCAAGAAAGTACCCCAAGTGTCAACTCCAACTCTTGTAGAGGTCTCAAGAAACCTAGGAAAAGTGGGCAGCAAATGTTGTAAACATCCTGAAGCAAAAAGAATGCCCTGTGCAGAAGACTATCTATCCGTGGTCCTGAACCAGTTATGTGTGTTGCATGAGAAAACGCCAGTAAGTGACAGAGTCACCAAATGCTGCACAGAATCCTTGGTGAACAGGCGACCATGCTTTTCAGCTCTGGAAGTCGATGAAACATACGTTCCCAAAGAGTTTAATGCTGAAACATTCACCTTCCATGCAGATATATGCACACTTTCTGAGAAGGAGAGACAAATCAAGAAACAAACTGCACTTGTTGAGCTTGTGAAACACAAGCCCAAGGCAACAAAAGAGCAACTGAAAGCTGTTATGGATGATTTCGCAGCTTTTGTAGAGAAGTGCTGCAAGGCTGACGATAAGGAGACCTGCTTTGCCGAGGAGGGTAAAAAACTTGTTGCTGCAAGTCGAGCTGCCTTAGGCTTATAACATCACATTTAAAAGCATCTCAGCCTACCATGAGAATAAGAGAAAGAAAATGAAGATCAAAAGCTTATTCATCTGTTTTTCTTTTTCGTTGGTGTAAAGCCA start: 27 end: 1965
Match: |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Subjc: TGGCACAATGAAGTGGGTAACCTTTATTTCCCTTCTTTTTCTCTTTAGCTCGGCTTATTCCAGGGGTGTGTTTCGTCGAGATGCACACAAGAGTGAGGTTGCTCATCGGTTTAAAGATTTGGGAGAAGAAAATTTCAAAGCCTTGGTGTTGATTGCCTTTGCTCAGTATCTTCAGCAGTGTCCATTTGAAGATCATGTAAAATTAGTGAATGAAGTAACTGAATTTGCAAAAACATGTGTTGCTGATGAGTCAGCTGAAAATTGTGACAAATCACTTCATACCCTTTTTGGAGACAAATTATGCACAGTTGCAACTCTTCGTGAAACCTATGGTGAAATGGCTGACTGCTGTGCAAAACAAGAACCTGAGAGAAATGAATGCTTCTTGCAACACAAAGATGACAACCCAAACCTCCCCCGATTGGTGAGACCAGAGGTTGATGTGATGTGCACTGCTTTTCATGACAATGAAGAGACATTTTTGAAAAAATACTTATATGAAATTGCCAGAAGACATCCTTACTTTTATGCCCCGGAACTCCTTTTCTTTGCTAAAAGGTATAAAGCTGCTTTTACAGAATGTTGCCAAGCTGCTGATAAAGCTGCCTGCCTGTTGCCAAAGCTCGATGAACTTCGGGATGAAGGGAAGGCTTCGTCTGCCAAACAGAGACTCAAGTGTGCCAGTCTCCAAAAATTTGGAGAAAGAGCTTTCAAAGCATGGGCAGTAGCTCGCCTGAGCCAGAGATTTCCCAAAGCTGAGTTTGCAGAAGTTTCCAAGTTAGTGACAGATCTTACCAAAGTCCACACGGAATGCTGCCATGGAGATCTGCTTGAATGTGCTGATGACAGGGCGGACCTTGCCAAGTATATCTGTGAAAATCAAGATTCGATCTCCAGTAAACTGAAGGAATGCTGTGAAAAACCTCTGTTGGAAAAATCCCACTGCATTGCCGAAGTGGAAAATGATGAGATGCCTGCTGACTTGCCTTCATTAGCTGCTGATTTTGTTGAAAGTAAGGATGTTTGCAAAAACTATGCTGAGGCAAAGGATGTCTTCCTGGGCATGTTTTTGTATGAATATGCAAGAAGGCATCCTGATTACTCTGTCGTGCTGCTGCTGAGACTTGCCAAGACATATGAAACCACTCTAGAGAAGTGCTGTGCCGCTGCAGATCCTCATGAATGCTATGCCAAAGTGTTCGATGAATTTAAACCTCTTGTGGAAGAGCCTCAGAATTTAATCAAACAAAATTGTGAGCTTTTTGAGCAGCTTGGAGAGTACAAATTCCAGAATGCGCTATTAGTTCGTTACACCAAGAAAGTACCCCAAGTGTCAACTCCAACTCTTGTAGAGGTCTCAAGAAACCTAGGAAAAGTGGGCAGCAAATGTTGTAAACATCCTGAAGCAAAAAGAATGCCCTGTGCAGAAGACTATCTATCCGTGGTCCTGAACCAGTTATGTGTGTTGCATGAGAAAACGCCAGTAAGTGACAGAGTCACCAAATGCTGCACAGAATCCTTGGTGAACAGGCGACCATGCTTTTCAGCTCTGGAAGTCGATGAAACATACGTTCCCAAAGAGTTTAATGCTGAAACATTCACCTTCCATGCAGATATATGCACACTTTCTGAGAAGGAGAGACAAATCAAGAAACAAACTGCACTTGTTGAGCTTGTGAAACACAAGCCCAAGGCAACAAAAGAGCAACTGAAAGCTGTTATGGATGATTTCGCAGCTTTTGTAGAGAAGTGCTGCAAGGCTGACGATAAGGAGACCTGCTTTGCCGAGGAGGGTAAAAAACTTGTTGCTGCAAGTCAAGCTGCCTTAGGCTTATAACATCACATTTAAAAGCATCTCAGCCTACCATGAGAATAAGAGAAAGAAAATGAAGATCAAAAGCTTATTCATCTGTTTTTCTTTTTCGTTGGTGTAAAGCCA start: 1 end: 1939
Align Title:gi|115607206|gb|DQ986150.1| Homo sapiens serum albumin mRNA, complete cds
Align Len: 1884
Score: 3733.0 Bits: 3367.27 E-val: 0.0
N.aligns:None Ident:1877 Pos.:1877 Gaps:0 Align len:1884
Strand: (None, None) Frame: (1, 1)
Query: CCTTTGGCACAATGAAGTGGGTAACCTTTATTTCCCTTCTTTTTCTCTTTAGCTCGGCTTATTCCAGGGGTGTGTTTCGTCGAGATGCACACAAGAGTGAGGTTGCTCATCGGTTTAAAGATTTGGGAGAAGAAAATTTCAAAGCCTTAGTGTTGATTGCCTTTGCTCAGTATCTTCAGCAGTGTCCATTTGAAGATCATGTAAAATTAGTGAATGAAGTAACTGAATTTGCAAAAACATGTGTTGCTGATGAGTCAGCTGAAAATTGTGACAAATCACTTCATACCCTTTTTGGAGACAAATTATGCACAGTTGCAACTCTTCGTGAAACCTATGGTGAAATGGCTGACTGCTGTGCAAAACAAGAACCTGAGAGAAATGAATGCTTCTTGCAACACAAAGATGACAACCCAAACCTCCCCCGATTGGTGAGACCAGAGGTTGATGTGATGTGCACTGCTTTTCATGACAATGAAGAGACATTTTTGAAAAAATACTTATATGAAATTGCCAGAAGACATCCTTACTTCTATGCCCCGGAACTCCTTTTCTTTGCTAAAAGGTATAAAGCTGCTTTTACAGAATGTTGCCAAGCTGCTGATAAGGCTGCCTGCCTGTTGCCAAAGCTCGATGAACTTCGGGATGAAGGGAAGGCTTCGTCTGCCAAACAGAGGCTCAAGTGTGCCAGTCTCCAAAAATTTGGAGAAAGAGCTTTCAAAGCATGGGCAGTAGCTCGCCTGAGCCAGAGATTTCCCAAAGCTGAGTTTGCAGAAGTTTCCAAGTTAGTGACAGATCTTACCAAAGTCCACACGGAATGCTGCCATGGAGATCTGCTTGAATGTGCTGATGACAGGGCGGACCTTGCCAAGTATATCTGTGAAAATCAAGATTCGATCTCCAGTAAACTGAAGGAATGCTGTGAAAAACCTCTGTTGGAAAAATCCCACTGCATTGCCGAAGTGGAAAATGATGAGATGCCTGCTGACTTGCCTTCATTAGCTGCTGATTTTGTTGAAAGTAAGGATGTTTGCAAAAACTATGCTGAGGCAAAGGATGTCTTCCTGGGCATGTTTTTGTATGAATATGCAAGAAGGCATCCTGATTACTCTGTCGTGCTGCTGCTGAGACTTGCCAAGACATATAAAACCACTCTAGAGAAGTGCTGTGCCGCTGCAGATCCTCATGAATGCTATGCCAAAGTGTTCGATGAATTTAAACCTCTTGTGGAAGAGCCTCAGAATTTAATCAAACAAAATTGTGAGCTTTTTGAGCAGCTTGGAGAGTACAAATTCCAGAATGCGCTATTAGTTCGTTACACCAAGAAAGTACCCCAAGTGTCAACTCCAACTCTTGTAGAGGTCTCAAGAAACCTAGGAAAAGTGGGCAGCAAATGTTGTAAACATCCTGAAGCAAAAAGAATGCCCTGTGCAGAAGACTATCTATCCGTGGTCCTGAACCAGTTATGTGTGTTGCATGAGAAAACGCCAGTAAGTGACAGAGTCACCAAATGCTGCACAGAATCCTTGGTGAACAGGCGACCATGCTTTTCAGCTCTGGAAGTCGATGAAACATACGTTCCCAAAGAGTTTAATGCTGAAACATTCACCTTCCATGCAGATATATGCACACTTTCTGAGAAGGAGAGACAAATCAAGAAACAAACTGCACTTGTTGAGCTTGTGAAACACAAGCCCAAGGCAACAAAAGAGCAACTGAAAGCTGTTATGGATGATTTCGCAGCTTTTGTAGAGAAGTGCTGCAAGGCTGACGATAAGGAGACCTGCTTTGCCGAGGAGGGTAAAAAACTTGTTGCTGCAAGTCGAGCTGCCTTAGGCTTATAACATCACATTTAAAAGCATCTCAGCCTACCATGAGAATAAGAGA start: 23 end: 1906
Match: |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Subjc: CCTTTGGCACAATGAAGTGGGTAACCTTTATTTCCCTTCTTTTTCTCTTTAGCTCGGCTTATTCCAGGGGTGTGTTTCGTCGAGATGCACACAAGAGTGAGGTTGCTCATCGGTTTAAAGATTTGGGAGAAGAAAATTTCAAAGCCTTGGTGTTGATTGCCTTTGCTCAGTATCTTCAGCAGTGTCCATTTGAAGATCATGTAAAATTAGTGAATGAAGTAACTGAATTTGCAAAAACATGTGTTGCTGATGAGTCAGCTGAAAATTGTGACAAATCACTTCATACCCTTTTTGGAGACAAATTATGCACAGTTGCAACTCTTCGTGAAACCTATGGTGAAATGGCTGACTGCTGTGCAAAACAAGAACCTGAGAGAAATGAATGCTTCTTGCAACACAAAGATGACAACCCAAACCTCCCCCGATTGGTGAGACCAGAGGTTGATGTGATGTGCACTGCTTTTCATGACAATGAAGAGACATTTTTGAAAAAATACTTATATGAAATTGCCAGAAGACATCCTTACTTTTATGCCCCGGAACTCCTTTTCTTTGCTAAAAGGTATAAAGCTGCTTTTACAGAATGTTGCCAAGCTGCTGATAAAGCTGCCTGCCTGTTGCCAAAGCTCGATGAACTTCGGGATGAAGGGAAGGCTTCGTCTGCCAAACAGAGACTCAAGTGTGCCAGTCTCCAAAAATTTGGAGAAAGAGCTTTCAAAGCATGGGCAGTAGCTCGCCTGAGCCAGAGATTTCCCAAAGCTGAGTTTGCAGAAGTTTCCAAGTTAGTGACAGATCTTACCAAAGTCCACACGGAATGCTGCCATGGAGATCTGCTTGAATGTGCTGATGACAGGGCGGACCTTGCCAAGTATATCTGTGAAAATCAAGATTCGATCTCCAGTAAACTGAAGGAATGCTGTGAAAAACCTCTGTTGGAAAAATCCCACTGCATTGCCGAAGTGGAAAATGATGAGATGCCTGCTGACTTGCCTTCATTAGCTGCTGATTTTGTTGAAAGTAAGGATGTTTGCAAAAACTATGCTGAGGCAAAGGATGTCTTCCTGGGCATGTTTTTGTATGAATATGCAAGAAGGCATCCTGATTACTCTGTCGTGCTGCTGCTGAGACTTGCCAAGACATATGAAACCACTCTAGAGAAGTGCTGTGCCGCTGCAGATCCTCATGAATGCTATGCCAAAGTGTTCGATGAATTTAAACCTCTTGTGGAAGAGCCTCAGAATTTAATCAAACAAAATTGTGAGCTTTTTGAGCAGCTTGGAGAGTACAAATTCCAGAATGCGCTATTAGTTCGTTACACCAAGAAAGTACCCCAAGTGTCAACTCCAACTCTTGTAGAGGTCTCAAGAAACCTAGGAAAAGTGGGCAGCAAATGTTGTAAACATCCTGAAGCAAAAAGAATGCCCTGTGCAGAAGACTATCTATCCGTGGTCCTGAACCAGTTATGTGTGTTGCATGAGAAAACGCCAGTAAGTGACAGAGTCACCAAATGCTGCACAGAATCCTTGGTGAACAGGCGACCATGCTTTTCAGCTCTGGAAGTCGATGAAACATACGTTCCCAAAGAGTTTAATGCTGAAACATTCACCTTCCATGCAGATATATGCACACTTTCTGAGAAGGAGAGACAAATCAAGAAACAAACTGCACTTGTTGAGCTCGTGAAACACAAGCCCAAGGCAACAAAAGAGCAACTGAAAGCTGTTATGGATGATTTCGCAGCTTTTGTAGAGAAGTGCTGCAAGGCTGACGATAAGGAGACCTGCTTTGCCGAGGAGGGTAAAAAACTTGTTGCTGCAAGTCAAGCTGCCTTAGGCTTATAACATCACATTTAAAAGCATCTCAGCCTACCATGAGAATAAGAGA start: 1 end: 1884
Align Title:gi|164692402|dbj|AK314794.1| Homo sapiens cDNA, FLJ95666, highly similar to Homo sapiens albumin (ALB), mRNA
Align Len: 1871
Score: 3676.0 Bits: 3315.88 E-val: 0.0
N.aligns:None Ident:1853 Pos.:1853 Gaps:0 Align len:1863
Strand: (None, None) Frame: (1, 1)
Query: TCTCTTCTGTCAACCCCACGCGCCTTTGGCACAATGAAGTGGGTAACCTTTATTTCCCTTCTTTTTCTCTTTAGCTCGGCTTATTCCAGGGGTGTGTTTCGTCGAGATGCACACAAGAGTGAGGTTGCTCATCGGTTTAAAGATTTGGGAGAAGAAAATTTCAAAGCCTTAGTGTTGATTGCCTTTGCTCAGTATCTTCAGCAGTGTCCATTTGAAGATCATGTAAAATTAGTGAATGAAGTAACTGAATTTGCAAAAACATGTGTTGCTGATGAGTCAGCTGAAAATTGTGACAAATCACTTCATACCCTTTTTGGAGACAAATTATGCACAGTTGCAACTCTTCGTGAAACCTATGGTGAAATGGCTGACTGCTGTGCAAAACAAGAACCTGAGAGAAATGAATGCTTCTTGCAACACAAAGATGACAACCCAAACCTCCCCCGATTGGTGAGACCAGAGGTTGATGTGATGTGCACTGCTTTTCATGACAATGAAGAGACATTTTTGAAAAAATACTTATATGAAATTGCCAGAAGACATCCTTACTTCTATGCCCCGGAACTCCTTTTCTTTGCTAAAAGGTATAAAGCTGCTTTTACAGAATGTTGCCAAGCTGCTGATAAGGCTGCCTGCCTGTTGCCAAAGCTCGATGAACTTCGGGATGAAGGGAAGGCTTCGTCTGCCAAACAGAGGCTCAAGTGTGCCAGTCTCCAAAAATTTGGAGAAAGAGCTTTCAAAGCATGGGCAGTAGCTCGCCTGAGCCAGAGATTTCCCAAAGCTGAGTTTGCAGAAGTTTCCAAGTTAGTGACAGATCTTACCAAAGTCCACACGGAATGCTGCCATGGAGATCTGCTTGAATGTGCTGATGACAGGGCGGACCTTGCCAAGTATATCTGTGAAAATCAAGATTCGATCTCCAGTAAACTGAAGGAATGCTGTGAAAAACCTCTGTTGGAAAAATCCCACTGCATTGCCGAAGTGGAAAATGATGAGATGCCTGCTGACTTGCCTTCATTAGCTGCTGATTTTGTTGAAAGTAAGGATGTTTGCAAAAACTATGCTGAGGCAAAGGATGTCTTCCTGGGCATGTTTTTGTATGAATATGCAAGAAGGCATCCTGATTACTCTGTCGTGCTGCTGCTGAGACTTGCCAAGACATATAAAACCACTCTAGAGAAGTGCTGTGCCGCTGCAGATCCTCATGAATGCTATGCCAAAGTGTTCGATGAATTTAAACCTCTTGTGGAAGAGCCTCAGAATTTAATCAAACAAAATTGTGAGCTTTTTGAGCAGCTTGGAGAGTACAAATTCCAGAATGCGCTATTAGTTCGTTACACCAAGAAAGTACCCCAAGTGTCAACTCCAACTCTTGTAGAGGTCTCAAGAAACCTAGGAAAAGTGGGCAGCAAATGTTGTAAACATCCTGAAGCAAAAAGAATGCCCTGTGCAGAAGACTATCTATCCGTGGTCCTGAACCAGTTATGTGTGTTGCATGAGAAAACGCCAGTAAGTGACAGAGTCACCAAATGCTGCACAGAATCCTTGGTGAACAGGCGACCATGCTTTTCAGCTCTGGAAGTCGATGAAACATACGTTCCCAAAGAGTTTAATGCTGAAACATTCACCTTCCATGCAGATATATGCACACTTTCTGAGAAGGAGAGACAAATCAAGAAACAAACTGCACTTGTTGAGCTTGTGAAACACAAGCCCAAGGCAACAAAAGAGCAACTGAAAGCTGTTATGGATGATTTCGCAGCTTTTGTAGAGAAGTGCTGCAAGGCTGACGATAAGGAGACCTGCTTTGCCGAGGAGGGTAAAAAACTTGTTGCTGCAAGTCGAGCTGCCTTAGGCTTATAA start: 1 end: 1863
Match: ||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||
Subjc: TCTCTTCTGTCAACCCCACACGCCTTTGGCACAATGAAGTGGGTAACCTTTATTTCCCTTCTTTTTCTCTTTAGCTCGGCTTATTCCAGGGGTGTGTTTCGTCGAGATGCACACAAGAGTGAGGTTGCTCATCGGTTTAAAGATTTGGGAGAAGAAAATTTCAAAGCCTTGGTGTTGATTGCCTTTGCTCAGTATCTTCAGCAGTGTCCATTTGAAGATCATGTAAAATTAGTGAATGAAGTAACTGAATTTGCAAAAACATGTGTTGCTGATGAGTCAGCTGAAAATTGTGACAAATCACTTCATACCCTTTTTGGAGACAAATTATGCACAGTTGCAACTCTTCGTGAAACCTATGGTGAAATGGCTGACTGCTGTGCAAAACAAGAACCTGAGAGAAATGAATGCTTCTTGCAACACAAAGATGACAACCCAAACCTCCCCCGATTGGTGAGACCAGAGGTTGATGTGATGTGCACTGCTTTTCATGACAATGAAGAGACATTTTTGAAAAAATACTTATATGAAATTGCCAGAAGACATCCTTACTTTTATGCCCCGGAACTCCTTTTCTTTGCTAAAAGGTATAAAGCTGCTTTTACAGAATGTTGCCAAGCTGCTGATAAAGCTGCCTGCCTGTTGCCAAAGCTCGATGAACTTCGGGATGAAGGGAAGGCTTCGTCTGCCAAACAGAGACTCAAGTGTGCCAGTCTCCAAAAATTTGGAGAAAGAGCTTTCAAAGCATGGGCAGTAGCTCGCCTGAGCCAGAGATTTCCCAAAGCTGAGTTTGCAGAAGTTTCCAAGTTAGTGACAGATCTTACCAAAGTCCACACGGAATGCTGCCATGGAGATCTGCTTGAATGTGCTGATGACAGGGCGGACCTTGCCAAGTATATCTGTGAAAATCAAGATTCGATCTCCAGTAAACTGAAGGAATGCTGTGAAAAACCTCTGTTGGAAAAATCCCACTGCATTGCCGAAGTGGAAAATGATGAGATGCCTGCTGACTTGCCTTCATTAGCTGCTGATTTTGTTGAAAGTAAGGATGTTTGCAAAAACTATGCTGAGGCAAAGGATGTCCTCCTGGGCATGTTTTTGTATGAATATGCAAGAAGGCATCCTGATTACTTTGTCGTGCTGCTGCTGAGACTTGCCAAGACATATGAAACCACTCTAGAGAAGTGCTGTGCCGCTGCAGATCCTCATGAATGCTATGCCAAAGTGTTCGATGAATTTAAACCTCTTGTGGAAGAGCCTCAGAATTTAATCAAACAAAATTGTGAGCTTTTTGAGCAGCTTGGAGAGTACAAATTCCAGAATGCGCTATTAGTTCGTTACACCAAGAAAGTACCCCAAGTGTCAACTCCAACTCTTGTAGAGGTCTCAAGAAACCTAGGAAAAGTGGGCAGCAAATGTTGTAAACATCCTGAAGCAAAAAGAATGCCCTGTGCAGAAGACTATCTATCCGTGGTCCTGAACCAGTTATGTGTGTTGCATGAGAAAACGCCAGTAAGTGACAGAGTCACCAAATGCTGCACAGAATCCTTGGTGAATAGGCGACCATGCTTTTCAGCTCTGGAAGTCGATGAAACATACGTTCCCAAAGAGTTTAATGCTGAAACATTCACCTTCCATGCAGATATATGCACACTTTCTGAGAAGGAGAGACAAATCAAGAAACAAACTGCACTTGTTGAGCTTGTGAAACACAAGCCCAAGGCAACAAAAGAGCAACTGAAAGCTGTTATGGATGATTTCGCAGCTTTTGTAGAGAAGTGCTGCAAGGCTGACGATAAGGAGACCTGCTTTGCCGAGGAGGGTAAAAAACTTGTTGCTGCAAGTCAAGCTGCCTTAGGCTTATAA start: 9 end: 1871
Align Title:gi|52001696|gb|AY728024.1| Homo sapiens serum albumin precursor, mRNA, complete cds
Align Len: 1842
Score: 3630.0 Bits: 3274.4 E-val: 0.0
N.aligns:None Ident:1824 Pos.:1824 Gaps:0 Align len:1830
Strand: (None, None) Frame: (1, 1)
Query: ATGAAGTGGGTAACCTTTATTTCCCTTCTTTTTCTCTTTAGCTCGGCTTATTCCAGGGGTGTGTTTCGTCGAGATGCACACAAGAGTGAGGTTGCTCATCGGTTTAAAGATTTGGGAGAAGAAAATTTCAAAGCCTTAGTGTTGATTGCCTTTGCTCAGTATCTTCAGCAGTGTCCATTTGAAGATCATGTAAAATTAGTGAATGAAGTAACTGAATTTGCAAAAACATGTGTTGCTGATGAGTCAGCTGAAAATTGTGACAAATCACTTCATACCCTTTTTGGAGACAAATTATGCACAGTTGCAACTCTTCGTGAAACCTATGGTGAAATGGCTGACTGCTGTGCAAAACAAGAACCTGAGAGAAATGAATGCTTCTTGCAACACAAAGATGACAACCCAAACCTCCCCCGATTGGTGAGACCAGAGGTTGATGTGATGTGCACTGCTTTTCATGACAATGAAGAGACATTTTTGAAAAAATACTTATATGAAATTGCCAGAAGACATCCTTACTTCTATGCCCCGGAACTCCTTTTCTTTGCTAAAAGGTATAAAGCTGCTTTTACAGAATGTTGCCAAGCTGCTGATAAGGCTGCCTGCCTGTTGCCAAAGCTCGATGAACTTCGGGATGAAGGGAAGGCTTCGTCTGCCAAACAGAGGCTCAAGTGTGCCAGTCTCCAAAAATTTGGAGAAAGAGCTTTCAAAGCATGGGCAGTAGCTCGCCTGAGCCAGAGATTTCCCAAAGCTGAGTTTGCAGAAGTTTCCAAGTTAGTGACAGATCTTACCAAAGTCCACACGGAATGCTGCCATGGAGATCTGCTTGAATGTGCTGATGACAGGGCGGACCTTGCCAAGTATATCTGTGAAAATCAAGATTCGATCTCCAGTAAACTGAAGGAATGCTGTGAAAAACCTCTGTTGGAAAAATCCCACTGCATTGCCGAAGTGGAAAATGATGAGATGCCTGCTGACTTGCCTTCATTAGCTGCTGATTTTGTTGAAAGTAAGGATGTTTGCAAAAACTATGCTGAGGCAAAGGATGTCTTCCTGGGCATGTTTTTGTATGAATATGCAAGAAGGCATCCTGATTACTCTGTCGTGCTGCTGCTGAGACTTGCCAAGACATATAAAACCACTCTAGAGAAGTGCTGTGCCGCTGCAGATCCTCATGAATGCTATGCCAAAGTGTTCGATGAATTTAAACCTCTTGTGGAAGAGCCTCAGAATTTAATCAAACAAAATTGTGAGCTTTTTGAGCAGCTTGGAGAGTACAAATTCCAGAATGCGCTATTAGTTCGTTACACCAAGAAAGTACCCCAAGTGTCAACTCCAACTCTTGTAGAGGTCTCAAGAAACCTAGGAAAAGTGGGCAGCAAATGTTGTAAACATCCTGAAGCAAAAAGAATGCCCTGTGCAGAAGACTATCTATCCGTGGTCCTGAACCAGTTATGTGTGTTGCATGAGAAAACGCCAGTAAGTGACAGAGTCACCAAATGCTGCACAGAATCCTTGGTGAACAGGCGACCATGCTTTTCAGCTCTGGAAGTCGATGAAACATACGTTCCCAAAGAGTTTAATGCTGAAACATTCACCTTCCATGCAGATATATGCACACTTTCTGAGAAGGAGAGACAAATCAAGAAACAAACTGCACTTGTTGAGCTTGTGAAACACAAGCCCAAGGCAACAAAAGAGCAACTGAAAGCTGTTATGGATGATTTCGCAGCTTTTGTAGAGAAGTGCTGCAAGGCTGACGATAAGGAGACCTGCTTTGCCGAGGAGGGTAAAAAACTTGTTGCTGCAAGTCGAGCTGCCTTAGGCTTATAA start: 34 end: 1863
Match: ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||
Subjc: ATGAAGTGGGTAACCTTTATTTCCCTTCTTTTTCTCTTTAGCTCGGCTTATTCCAGGGGTGTGTTTCGTCGAGATGCACACAAGAGTGAGGTTGCTCATCGGTTTAAAGATTTGGGAGAAGAAAATTTCAAAGCCTTGGTGTTGATTGCCTTTGCTCAGTATCTTCAGCAGTGTCCATTTGAAGATCATGTAAAATTAGTGAATGAAGTAACTGAATTTGCAAAAACATGTGTTGCTGATGAGTCAGCTGAAAATTGTGACAAATCACTTCATACCCTTTTTGGAGACAAATTATGCACAGTTGCAACTCTTCGTGAAACCTATGGTGAAATGGCTGACTGCTGTGCAAAACAAGAACCTGAGAGAAATGAATGCTTCTTGCAACACAAAGATGACAACCCAAACCTCCCCCGATTGGTGAGACCAGAGGTTGATGTGATGTGCACTGCTTTTCATGACAATGAAGAGACATTTTTGAAAAAATACTTATATGAAATTGCCAGAAGACATCCTTACTTTTATGCCCCGGAACTCCTTTTCTTTGCTAAAAGGTATAAAGCTGCTTTTACAGAATGTTGCCAAGCTGCTGATAAAGCTGCCTGCCTGTTGCCAAAGCTCGATGAACTTCGGGATGAAGGGAAGGCTTCGTCTGCCAAACAGAGACTCAAGTGTGCCAGTCTCCAAAAATTTGGAGAAAGAGCTTTCAAAGCATGGGCAGTAGCTCGCCTGAGCCAGAGATTTCCCAAAGCTGAGTTTGCAGAAGTTTCCAAGTTAGTGACAGATCTTACCAAAGTCCACACGGAATGCTGCCATGGAGATCTGCTTGAATGTGCTGATGACAGGGCGGACCTTGCCAAGTATATCTGTGAAAATCAAGATTCGATCTCCAGTAAACTGAAGGAATGCTGTGAAAAACCTCTGTTGGAAAAATCCCACTGCATTGCCGAAGTGGAAAATGATGAGATGCCTGCTGACTTGCCTTCATTAGCTGCTGATTTTGTTGAAAGTAAGGATGTTTGCAAAAACTATGCTGAGGCAAAGGATGTCTTCCTGGGCATGTTTTTGTATGAATATGCAAGAAGGCATCCTGATTACTCTGTCGTGCTGCTGCTGAGACTTGCCAAGACATATGAAACCACTCTAGAGAAGTGCTGTGCCGCTGCAGATCCTCATGAATGCTATGCCAAAGTGTTCGATGAATTTAAACCTCTTGTGGAAGAGCCTCAGAATTTAATCAAACAAAATTGTGAGCTTTTTGAGCAGCTTGGAGAGTACAAATTCCAGAATGCGCTATTAGTTCGTTACACCAAGAAAGTACCCCAAGTGTCAACTCCAACTCTTGTAGAGGTCTCAAGAAACCTAGGAAAAGTGGGCAGCAAATGTTGTAAACATCCTGAAGCAAAAAGAATGCCCTGTGCAGAAGACTATCTATCCGTGGTCCTGAACCAGTTATGTGTGTTGCATGAGAAAACGCCAGTAAGTGACAGAGTCACCAAATGCTGCACAGAATCCTTGGTGAACAGGCGACCATGCTTTTCAGCTCTGGAAGTCGATGAAACATACGTTCCCAAAGAGTTTAATGCTGAAACATTCACCTTCCATGCAGATATATGCACACTTTCTGAGAAGGAGAGACAAATCAAGAAACAAACTGCACTTGTTGAGCTTGTGAAACACAAGCCCAAGGCAACAAAAGAGCAACTGAAAGCTGTTATGGATGATTTCGCAGCTTTTGTAGAGAAGTGCTGCAAGGCTGACGATAAGGAGACCTGCTTTGCCGAGGAGGGTAAAAAACTTGTTGCTGCAAGTCAAGCTGCCTTAGGCTTATAA start: 7 end: 1836
Align Title:gi|123981021|gb|DQ891414.2| Synthetic construct clone IMAGE:100004044; FLH176691.01X; RZPDo839G03122D albumin (ALB) gene, encodes complete protein
Align Len: 1870
Score: 3626.0 Bits: 3270.79 E-val: 0.0
N.aligns:None Ident:1825 Pos.:1825 Gaps:0 Align len:1833
Strand: (None, None) Frame: (1, 1)
Query: CACAATGAAGTGGGTAACCTTTATTTCCCTTCTTTTTCTCTTTAGCTCGGCTTATTCCAGGGGTGTGTTTCGTCGAGATGCACACAAGAGTGAGGTTGCTCATCGGTTTAAAGATTTGGGAGAAGAAAATTTCAAAGCCTTAGTGTTGATTGCCTTTGCTCAGTATCTTCAGCAGTGTCCATTTGAAGATCATGTAAAATTAGTGAATGAAGTAACTGAATTTGCAAAAACATGTGTTGCTGATGAGTCAGCTGAAAATTGTGACAAATCACTTCATACCCTTTTTGGAGACAAATTATGCACAGTTGCAACTCTTCGTGAAACCTATGGTGAAATGGCTGACTGCTGTGCAAAACAAGAACCTGAGAGAAATGAATGCTTCTTGCAACACAAAGATGACAACCCAAACCTCCCCCGATTGGTGAGACCAGAGGTTGATGTGATGTGCACTGCTTTTCATGACAATGAAGAGACATTTTTGAAAAAATACTTATATGAAATTGCCAGAAGACATCCTTACTTCTATGCCCCGGAACTCCTTTTCTTTGCTAAAAGGTATAAAGCTGCTTTTACAGAATGTTGCCAAGCTGCTGATAAGGCTGCCTGCCTGTTGCCAAAGCTCGATGAACTTCGGGATGAAGGGAAGGCTTCGTCTGCCAAACAGAGGCTCAAGTGTGCCAGTCTCCAAAAATTTGGAGAAAGAGCTTTCAAAGCATGGGCAGTAGCTCGCCTGAGCCAGAGATTTCCCAAAGCTGAGTTTGCAGAAGTTTCCAAGTTAGTGACAGATCTTACCAAAGTCCACACGGAATGCTGCCATGGAGATCTGCTTGAATGTGCTGATGACAGGGCGGACCTTGCCAAGTATATCTGTGAAAATCAAGATTCGATCTCCAGTAAACTGAAGGAATGCTGTGAAAAACCTCTGTTGGAAAAATCCCACTGCATTGCCGAAGTGGAAAATGATGAGATGCCTGCTGACTTGCCTTCATTAGCTGCTGATTTTGTTGAAAGTAAGGATGTTTGCAAAAACTATGCTGAGGCAAAGGATGTCTTCCTGGGCATGTTTTTGTATGAATATGCAAGAAGGCATCCTGATTACTCTGTCGTGCTGCTGCTGAGACTTGCCAAGACATATAAAACCACTCTAGAGAAGTGCTGTGCCGCTGCAGATCCTCATGAATGCTATGCCAAAGTGTTCGATGAATTTAAACCTCTTGTGGAAGAGCCTCAGAATTTAATCAAACAAAATTGTGAGCTTTTTGAGCAGCTTGGAGAGTACAAATTCCAGAATGCGCTATTAGTTCGTTACACCAAGAAAGTACCCCAAGTGTCAACTCCAACTCTTGTAGAGGTCTCAAGAAACCTAGGAAAAGTGGGCAGCAAATGTTGTAAACATCCTGAAGCAAAAAGAATGCCCTGTGCAGAAGACTATCTATCCGTGGTCCTGAACCAGTTATGTGTGTTGCATGAGAAAACGCCAGTAAGTGACAGAGTCACCAAATGCTGCACAGAATCCTTGGTGAACAGGCGACCATGCTTTTCAGCTCTGGAAGTCGATGAAACATACGTTCCCAAAGAGTTTAATGCTGAAACATTCACCTTCCATGCAGATATATGCACACTTTCTGAGAAGGAGAGACAAATCAAGAAACAAACTGCACTTGTTGAGCTTGTGAAACACAAGCCCAAGGCAACAAAAGAGCAACTGAAAGCTGTTATGGATGATTTCGCAGCTTTTGTAGAGAAGTGCTGCAAGGCTGACGATAAGGAGACCTGCTTTGCCGAGGAGGGTAAAAAACTTGTTGCTGCAAGTCGAGCTGCCTTAGGCTTATA start: 30 end: 1862
Match: ||| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| ||||||||||||||||||
Subjc: CACCATGAAGTGGGTAACCTTTATTTCCCTTCTTTTTCTCTTTAGCTCGGCTTATTCCAGGGGTGTGTTTCGTCGAGATGCACACAAGAGTGAGGTTGCTCATCGGTTTAAAGATTTGGGAGAAGAAAATTTCAAAGCCTTGGTGTTGATTGCCTTTGCTCAGTATCTTCAGCAGTGTCCATTTGAAGATCATGTAAAATTAGTGAATGAAGTAACTGAATTTGCAAAAACATGTGTTGCTGATGAGTCAGCTGAAAATTGTGACAAATCACTTCATACCCTTTTTGGAGACAAATTATGCACAGTTGCAACTCTTCGTGAAACCTATGGTGAAATGGCTGACTGCTGTGCAAAACAAGAACCTGAGAGAAATGAATGCTTCTTGCAACACAAAGATGACAACCCAAACCTCCCCCGATTGGTGAGACCAGAGGTTGATGTGATGTGCACTGCTTTTCATGACAATGAAGAGACATTTTTGAAAAAATACTTATATGAAATTGCCAGAAGACATCCTTACTTTTATGCCCCGGAACTCCTTTTCTTTGCTAAAAGGTATAAAGCTGCTTTTACAGAATGTTGCCAAGCTGCTGATAAAGCTGCCTGCCTGTTGCCAAAGCTCGATGAACTTCGGGATGAAGGGAAGGCTTCGTCTGCCAAACAGAGACTCAAGTGTGCCAGTCTCCAAAAATTTGGAGAAAGAGCTTTCAAAGCATGGGCAGTAGCTCGCCTGAGCCAGAGATTTCCCAAAGCTGAGTTTGCAGAAGTTTCCAAGTTAGTGACAGATCTTACCAAAGTCCACACGGAATGCTGCCATGGAGATCTGCTTGAATGTGCTGATGACAGGGCGGACCTTGCCAAGTATATCTGTGAAAATCAAGATTCGATCTCCAGTAAACTGAAGGAATGCTGTGAAAAACCTCTGTTGGAAAAATCCCACTGCATTGCCGAAGTGGAAAATGATGAGATGCCTGCTGACTTGCCTTCATTAGCTGCTGATTTTGTTGAAAGTAAGGATGTTTGCAAAAACTATGCTGAGGCAAAGGATGTCTTCCTGGGCATGTTTTTGTATGAATATGCAAGAAGGCATCCTGATTACTCTGTCGTGCTGCTGCTGAGACTTGCCAAGACATATGAAACCACTCTAGAGAAGTGCTGTGCCGCTGCAGATCCTCATGAATGCTATGCCAAAGTGTTCGATGAATTTAAACCTCTTGTGGAAGAGCCTCAGAATTTAATCAAACAAAATTGTGAGCTTTTTGAGCAGCTTGGAGAGTACAAATTCCAGAATGCGCTATTAGTTCGTTACACCAAGAAAGTACCCCAAGTGTCAACTCCAACTCTTGTAGAGGTCTCAAGAAACCTAGGAAAAGTGGGCAGCAAATGTTGTAAACATCCTGAAGCAAAAAGAATGCCCTGTGCAGAAGACTATCTATCCGTGGTCCTGAACCAGTTATGTGTGTTGCATGAGAAAACGCCAGTAAGTGACAGAGTCACCAAATGCTGCACAGAATCCTTGGTGAACAGGCGACCATGCTTTTCAGCTCTGGAAGTCGATGAAACATACGTTCCCAAAGAGTTTAATGCTGAAACGTTCACCTTCCATGCAGATATATGCACACTTTCTGAGAAGGAGAGACAAATCAAGAAACAAACTGCACTTGTTGAGCTTGTGAAACACAAGCCCAAGGCAACAAAAGAGCAACTGAAAGCTGTTATGGATGATTTCGCAGCTTTTGTAGAGAAGTGCTGCAAGGCTGACGATAAGGAGACCTGCTTTGCCGAGGAGGGTAAAAAACTTGTTGCTGCAAGTCAAGCTGCCTTAGGCTTATA start: 19 end: 1851
Align Title:gi|123995824|gb|DQ894588.2| Synthetic construct Homo sapiens clone IMAGE:100009048; FLH176687.01L; RZPDo839G03121D albumin (ALB) gene, encodes complete protein
Align Len: 1870
Score: 3624.0 Bits: 3268.99 E-val: 0.0
N.aligns:None Ident:1824 Pos.:1824 Gaps:0 Align len:1832
Strand: (None, None) Frame: (1, 1)
Query: CACAATGAAGTGGGTAACCTTTATTTCCCTTCTTTTTCTCTTTAGCTCGGCTTATTCCAGGGGTGTGTTTCGTCGAGATGCACACAAGAGTGAGGTTGCTCATCGGTTTAAAGATTTGGGAGAAGAAAATTTCAAAGCCTTAGTGTTGATTGCCTTTGCTCAGTATCTTCAGCAGTGTCCATTTGAAGATCATGTAAAATTAGTGAATGAAGTAACTGAATTTGCAAAAACATGTGTTGCTGATGAGTCAGCTGAAAATTGTGACAAATCACTTCATACCCTTTTTGGAGACAAATTATGCACAGTTGCAACTCTTCGTGAAACCTATGGTGAAATGGCTGACTGCTGTGCAAAACAAGAACCTGAGAGAAATGAATGCTTCTTGCAACACAAAGATGACAACCCAAACCTCCCCCGATTGGTGAGACCAGAGGTTGATGTGATGTGCACTGCTTTTCATGACAATGAAGAGACATTTTTGAAAAAATACTTATATGAAATTGCCAGAAGACATCCTTACTTCTATGCCCCGGAACTCCTTTTCTTTGCTAAAAGGTATAAAGCTGCTTTTACAGAATGTTGCCAAGCTGCTGATAAGGCTGCCTGCCTGTTGCCAAAGCTCGATGAACTTCGGGATGAAGGGAAGGCTTCGTCTGCCAAACAGAGGCTCAAGTGTGCCAGTCTCCAAAAATTTGGAGAAAGAGCTTTCAAAGCATGGGCAGTAGCTCGCCTGAGCCAGAGATTTCCCAAAGCTGAGTTTGCAGAAGTTTCCAAGTTAGTGACAGATCTTACCAAAGTCCACACGGAATGCTGCCATGGAGATCTGCTTGAATGTGCTGATGACAGGGCGGACCTTGCCAAGTATATCTGTGAAAATCAAGATTCGATCTCCAGTAAACTGAAGGAATGCTGTGAAAAACCTCTGTTGGAAAAATCCCACTGCATTGCCGAAGTGGAAAATGATGAGATGCCTGCTGACTTGCCTTCATTAGCTGCTGATTTTGTTGAAAGTAAGGATGTTTGCAAAAACTATGCTGAGGCAAAGGATGTCTTCCTGGGCATGTTTTTGTATGAATATGCAAGAAGGCATCCTGATTACTCTGTCGTGCTGCTGCTGAGACTTGCCAAGACATATAAAACCACTCTAGAGAAGTGCTGTGCCGCTGCAGATCCTCATGAATGCTATGCCAAAGTGTTCGATGAATTTAAACCTCTTGTGGAAGAGCCTCAGAATTTAATCAAACAAAATTGTGAGCTTTTTGAGCAGCTTGGAGAGTACAAATTCCAGAATGCGCTATTAGTTCGTTACACCAAGAAAGTACCCCAAGTGTCAACTCCAACTCTTGTAGAGGTCTCAAGAAACCTAGGAAAAGTGGGCAGCAAATGTTGTAAACATCCTGAAGCAAAAAGAATGCCCTGTGCAGAAGACTATCTATCCGTGGTCCTGAACCAGTTATGTGTGTTGCATGAGAAAACGCCAGTAAGTGACAGAGTCACCAAATGCTGCACAGAATCCTTGGTGAACAGGCGACCATGCTTTTCAGCTCTGGAAGTCGATGAAACATACGTTCCCAAAGAGTTTAATGCTGAAACATTCACCTTCCATGCAGATATATGCACACTTTCTGAGAAGGAGAGACAAATCAAGAAACAAACTGCACTTGTTGAGCTTGTGAAACACAAGCCCAAGGCAACAAAAGAGCAACTGAAAGCTGTTATGGATGATTTCGCAGCTTTTGTAGAGAAGTGCTGCAAGGCTGACGATAAGGAGACCTGCTTTGCCGAGGAGGGTAAAAAACTTGTTGCTGCAAGTCGAGCTGCCTTAGGCTTAT start: 30 end: 1861
Match: ||| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||
Subjc: CACCATGAAGTGGGTAACCTTTATTTCCCTTCTTTTTCTCTTTAGCTCGGCTTATTCCAGGGGTGTGTTTCGTCGAGATGCACACAAGAGTGAGGTTGCTCATCGGTTTAAAGATTTGGGAGAAGAAAATTTCAAAGCCTTGGTGTTGATTGCCTTTGCTCAGTATCTTCAGCAGTGTCCATTTGAAGATCATGTAAAATTAGTGAATGAAGTAACTGAATTTGCAAAAACATGTGTTGCTGATGAGTCAGCTGAAAATTGTGACAAATCACTTCATACCCTTTTTGGAGACAAATTATGCACAGTTGCAACTCTTCGTGAAACCTATGGTGAAATGGCTGACTGCTGTGCAAAACAAGAACCTGAGAGAAATGAATGCTTCTTGCAACACAAAGATGACAACCCAAACCTCCCCCGATTGGTGAGACCAGAGGTTGATGTGATGTGCACTGCTTTTCATGACAATGAAGAGACATTTTTGAAAAAATACTTATATGAAATTGCCAGAAGACATCCTTACTTTTATGCCCCGGAACTCCTTTTCTTTGCTAAAAGGTATAAAGCTGCTTTTACAGAATGTTGCCAAGCTGCTGATAAAGCTGCCTGCCTGTTGCCAAAGCTCGATGAACTTCGGGATGAAGGGAAGGCTTCGTCTGCCAAACAGAGACTCAAGTGTGCCAGTCTCCAAAAATTTGGAGAAAGAGCTTTCAAAGCATGGGCAGTAGCTCGCCTGAGCCAGAGATTTCCCAAAGCTGAGTTTGCAGAAGTTTCCAAGTTAGTGACAGATCTTACCAAAGTCCACACGGAATGCTGCCATGGAGATCTGCTTGAATGTGCTGATGACAGGGCGGACCTTGCCAAGTATATCTGTGAAAATCAAGATTCGATCTCCAGTAAACTGAAGGAATGCTGTGAAAAACCTCTGTTGGAAAAATCCCACTGCATTGCCGAAGTGGAAAATGATGAGATGCCTGCTGACTTGCCTTCATTAGCTGCTGATTTTGTTGAAAGTAAGGATGTTTGCAAAAACTATGCTGAGGCAAAGGATGTCTTCCTGGGCATGTTTTTGTATGAATATGCAAGAAGGCATCCTGATTACTCTGTCGTGCTGCTGCTGAGACTTGCCAAGACATATGAAACCACTCTAGAGAAGTGCTGTGCCGCTGCAGATCCTCATGAATGCTATGCCAAAGTGTTCGATGAATTTAAACCTCTTGTGGAAGAGCCTCAGAATTTAATCAAACAAAATTGTGAGCTTTTTGAGCAGCTTGGAGAGTACAAATTCCAGAATGCGCTATTAGTTCGTTACACCAAGAAAGTACCCCAAGTGTCAACTCCAACTCTTGTAGAGGTCTCAAGAAACCTAGGAAAAGTGGGCAGCAAATGTTGTAAACATCCTGAAGCAAAAAGAATGCCCTGTGCAGAAGACTATCTATCCGTGGTCCTGAACCAGTTATGTGTGTTGCATGAGAAAACGCCAGTAAGTGACAGAGTCACCAAATGCTGCACAGAATCCTTGGTGAACAGGCGACCATGCTTTTCAGCTCTGGAAGTCGATGAAACATACGTTCCCAAAGAGTTTAATGCTGAAACGTTCACCTTCCATGCAGATATATGCACACTTTCTGAGAAGGAGAGACAAATCAAGAAACAAACTGCACTTGTTGAGCTTGTGAAACACAAGCCCAAGGCAACAAAAGAGCAACTGAAAGCTGTTATGGATGATTTCGCAGCTTTTGTAGAGAAGTGCTGCAAGGCTGACGATAAGGAGACCTGCTTTGCCGAGGAGGGTAAAAAACTTGTTGCTGCAAGTCAAGCTGCCTTAGGCTTAT start: 19 end: 1850
Align Title:gi|6013426|gb|AF190168.1|AF190168 Homo sapiens serum albumin precursor, mRNA, complete cds
Align Len: 1830
Score: 3615.0 Bits: 3260.88 E-val: 0.0
N.aligns:None Ident:1821 Pos.:1821 Gaps:0 Align len:1830
Strand: (None, None) Frame: (1, 1)
Query: ATGAAGTGGGTAACCTTTATTTCCCTTCTTTTTCTCTTTAGCTCGGCTTATTCCAGGGGTGTGTTTCGTCGAGATGCACACAAGAGTGAGGTTGCTCATCGGTTTAAAGATTTGGGAGAAGAAAATTTCAAAGCCTTAGTGTTGATTGCCTTTGCTCAGTATCTTCAGCAGTGTCCATTTGAAGATCATGTAAAATTAGTGAATGAAGTAACTGAATTTGCAAAAACATGTGTTGCTGATGAGTCAGCTGAAAATTGTGACAAATCACTTCATACCCTTTTTGGAGACAAATTATGCACAGTTGCAACTCTTCGTGAAACCTATGGTGAAATGGCTGACTGCTGTGCAAAACAAGAACCTGAGAGAAATGAATGCTTCTTGCAACACAAAGATGACAACCCAAACCTCCCCCGATTGGTGAGACCAGAGGTTGATGTGATGTGCACTGCTTTTCATGACAATGAAGAGACATTTTTGAAAAAATACTTATATGAAATTGCCAGAAGACATCCTTACTTCTATGCCCCGGAACTCCTTTTCTTTGCTAAAAGGTATAAAGCTGCTTTTACAGAATGTTGCCAAGCTGCTGATAAGGCTGCCTGCCTGTTGCCAAAGCTCGATGAACTTCGGGATGAAGGGAAGGCTTCGTCTGCCAAACAGAGGCTCAAGTGTGCCAGTCTCCAAAAATTTGGAGAAAGAGCTTTCAAAGCATGGGCAGTAGCTCGCCTGAGCCAGAGATTTCCCAAAGCTGAGTTTGCAGAAGTTTCCAAGTTAGTGACAGATCTTACCAAAGTCCACACGGAATGCTGCCATGGAGATCTGCTTGAATGTGCTGATGACAGGGCGGACCTTGCCAAGTATATCTGTGAAAATCAAGATTCGATCTCCAGTAAACTGAAGGAATGCTGTGAAAAACCTCTGTTGGAAAAATCCCACTGCATTGCCGAAGTGGAAAATGATGAGATGCCTGCTGACTTGCCTTCATTAGCTGCTGATTTTGTTGAAAGTAAGGATGTTTGCAAAAACTATGCTGAGGCAAAGGATGTCTTCCTGGGCATGTTTTTGTATGAATATGCAAGAAGGCATCCTGATTACTCTGTCGTGCTGCTGCTGAGACTTGCCAAGACATATAAAACCACTCTAGAGAAGTGCTGTGCCGCTGCAGATCCTCATGAATGCTATGCCAAAGTGTTCGATGAATTTAAACCTCTTGTGGAAGAGCCTCAGAATTTAATCAAACAAAATTGTGAGCTTTTTGAGCAGCTTGGAGAGTACAAATTCCAGAATGCGCTATTAGTTCGTTACACCAAGAAAGTACCCCAAGTGTCAACTCCAACTCTTGTAGAGGTCTCAAGAAACCTAGGAAAAGTGGGCAGCAAATGTTGTAAACATCCTGAAGCAAAAAGAATGCCCTGTGCAGAAGACTATCTATCCGTGGTCCTGAACCAGTTATGTGTGTTGCATGAGAAAACGCCAGTAAGTGACAGAGTCACCAAATGCTGCACAGAATCCTTGGTGAACAGGCGACCATGCTTTTCAGCTCTGGAAGTCGATGAAACATACGTTCCCAAAGAGTTTAATGCTGAAACATTCACCTTCCATGCAGATATATGCACACTTTCTGAGAAGGAGAGACAAATCAAGAAACAAACTGCACTTGTTGAGCTTGTGAAACACAAGCCCAAGGCAACAAAAGAGCAACTGAAAGCTGTTATGGATGATTTCGCAGCTTTTGTAGAGAAGTGCTGCAAGGCTGACGATAAGGAGACCTGCTTTGCCGAGGAGGGTAAAAAACTTGTTGCTGCAAGTCGAGCTGCCTTAGGCTTATAA start: 34 end: 1863
Match: ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| ||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||
Subjc: ATGAAGTGGGTAACCTTTATTTCCCTTCTTTTTCTCTTTAGCTCGGCTTATTCCAGGGGTGTGTTTCGTCGAGATGCACACAAGAGTGAGGTTGCTCATCGGTTTAAAGATTTGGGAGAAGAAAATTTCAAAGCCTTGGTGTTGATTGCCTTTGCTCAGTATCTTCAGCAGTGTCCATTTGAAGATCATGTAAAATTAGTGAATGAAGTAACTGAATTTGCAAAAACATGTGTTGCTGATGAGTCAGCTGAAAATTGTGACAAATCACTTCATACCCTTTTTGGAGACAAATTATGCACAGTTGCAACTCTTCGTGAAACCTATGGTGAAATGGCTGACTGCTGTGCAAAACAAGAACCTGAGAGTAATGAATGCTTCTTGCAACACAAAGATGACAACCCAAACCTCCCCCGATTGGTGAGACCAGAGGTTGATGTGATGTGCACTGCTTTTCATGACAATGAAGAGACATTTTTGAAAAAATACCTATATGAAATTGCCAGAAGACATCCTTACTTTTATGCCCCGGAACTCCTTTTCTTTGCTAAAAGGTATAAAGCTGCTTTTACAGAATGTTGCCAAGCTGCTGATAAAGCTGCCTGCCTGTTGCCAAAGCTCGATGAACTTCGGGATGAAGGGAAGGCTTCGTCTGCCAAACAGAGACTCAAGTGTGCCAGTCTCCAAAAATTTGGAGAAAGAGCTTTCAAAGCATGGGCAGTAGCTCGCCTGAGCCAGAGATTTCCCAAAGCTGAGTTTGCAGAAGTTTCCAAGTTAGTGACAGATCTTACCAAAGTCCACACGGAATGCTGCCATGGAGATCTGCTTGAATGTGCTGATGACAGGGCGGACCTTGCCAAGTATATCTGTGAAAATCAAGATTCGATCTCCAGTAAACTGAAGGAATGCTGTGAAAAACCTCTGTTGGAAAAATCCCACTGCATTGCCGAAGTGGAAAATGATGAGATGCCTGCTGACTTGCCTTCATTAGCTGCTGATTTTGTTGAAAGTAAGGATGTTTGCAAAAACTATGCTGAGGCAAAGGATGTCTTCCTGGGCATGTTTTTGTATGAATATGCAAGAAGGCATCCTGATTACTCTGTCGTGCTGCTGCTGAGACTTGCCAAGACATATGAAACCACTCTAGAGAAGTGCTGTGCCGCTGCAGATCCTCATGAATGCTATGCCAAAGTGTTCGATGAATTTAAACCTCTTGTGGAAGAGCCTCAGAATTTAATCAAACAAAATTGTGAGCTTTTTGAGCAGCTTGGAGAGTACAAATTCCAGAATGCGCTATTAGTTCGTTACACCAAGAAAGTACCCCAAGTGTCAACTCCAACTCTTGTAGAGGTCTCAAGAAACCTAGGAAAAGTGGGCAGCAAATGTTGTAAACATCCTGGAGCAAAAAGAATGCCCTGTGCAGAAGACTATCTATCCGTGGTCCTGAACCAGTTATGTGTGTTGCATGAGAAAACGCCAGTAAGTGACAGAGTCACCAAATGCTGCACAGAATCCTTGGTGAACAGGCGACCATGCTTTTCAGCTCTGGAAGTCGATGAAACATACGTTCCCAAAGAGTTTAATGCTGAAACATTCACCTTCCATGCAGATATATGCACACTTTCTGAGAAGGAGAGACAAATCAAGAAACAAACTGCACTTGTTGAGCTTGTGAAACACAAGCCCAAGGCAACAAAAGAGCAACTGAAAGCTGTTATGGATGATTTCGCAGCTTTTGTAGAGAAGTGCTGCAAGGCTGACGATAAGGAGACCTGCTTTGCCGAGGAGGGTAAAAAACTTGTTGCTGCAAGTCAAGCTGCCTTAGGCTTATAA start: 1 end: 1830
Align Title:gi|332356379|gb|HQ537426.1| Homo sapiens albumin mRNA, partial cds
Align Len: 1758
Score: 3471.0 Bits: 3131.03 E-val: 0.0
N.aligns:None Ident:1749 Pos.:1749 Gaps:0 Align len:1758
Strand: (None, None) Frame: (1, 1)
Query: GATGCACACAAGAGTGAGGTTGCTCATCGGTTTAAAGATTTGGGAGAAGAAAATTTCAAAGCCTTAGTGTTGATTGCCTTTGCTCAGTATCTTCAGCAGTGTCCATTTGAAGATCATGTAAAATTAGTGAATGAAGTAACTGAATTTGCAAAAACATGTGTTGCTGATGAGTCAGCTGAAAATTGTGACAAATCACTTCATACCCTTTTTGGAGACAAATTATGCACAGTTGCAACTCTTCGTGAAACCTATGGTGAAATGGCTGACTGCTGTGCAAAACAAGAACCTGAGAGAAATGAATGCTTCTTGCAACACAAAGATGACAACCCAAACCTCCCCCGATTGGTGAGACCAGAGGTTGATGTGATGTGCACTGCTTTTCATGACAATGAAGAGACATTTTTGAAAAAATACTTATATGAAATTGCCAGAAGACATCCTTACTTCTATGCCCCGGAACTCCTTTTCTTTGCTAAAAGGTATAAAGCTGCTTTTACAGAATGTTGCCAAGCTGCTGATAAGGCTGCCTGCCTGTTGCCAAAGCTCGATGAACTTCGGGATGAAGGGAAGGCTTCGTCTGCCAAACAGAGGCTCAAGTGTGCCAGTCTCCAAAAATTTGGAGAAAGAGCTTTCAAAGCATGGGCAGTAGCTCGCCTGAGCCAGAGATTTCCCAAAGCTGAGTTTGCAGAAGTTTCCAAGTTAGTGACAGATCTTACCAAAGTCCACACGGAATGCTGCCATGGAGATCTGCTTGAATGTGCTGATGACAGGGCGGACCTTGCCAAGTATATCTGTGAAAATCAAGATTCGATCTCCAGTAAACTGAAGGAATGCTGTGAAAAACCTCTGTTGGAAAAATCCCACTGCATTGCCGAAGTGGAAAATGATGAGATGCCTGCTGACTTGCCTTCATTAGCTGCTGATTTTGTTGAAAGTAAGGATGTTTGCAAAAACTATGCTGAGGCAAAGGATGTCTTCCTGGGCATGTTTTTGTATGAATATGCAAGAAGGCATCCTGATTACTCTGTCGTGCTGCTGCTGAGACTTGCCAAGACATATAAAACCACTCTAGAGAAGTGCTGTGCCGCTGCAGATCCTCATGAATGCTATGCCAAAGTGTTCGATGAATTTAAACCTCTTGTGGAAGAGCCTCAGAATTTAATCAAACAAAATTGTGAGCTTTTTGAGCAGCTTGGAGAGTACAAATTCCAGAATGCGCTATTAGTTCGTTACACCAAGAAAGTACCCCAAGTGTCAACTCCAACTCTTGTAGAGGTCTCAAGAAACCTAGGAAAAGTGGGCAGCAAATGTTGTAAACATCCTGAAGCAAAAAGAATGCCCTGTGCAGAAGACTATCTATCCGTGGTCCTGAACCAGTTATGTGTGTTGCATGAGAAAACGCCAGTAAGTGACAGAGTCACCAAATGCTGCACAGAATCCTTGGTGAACAGGCGACCATGCTTTTCAGCTCTGGAAGTCGATGAAACATACGTTCCCAAAGAGTTTAATGCTGAAACATTCACCTTCCATGCAGATATATGCACACTTTCTGAGAAGGAGAGACAAATCAAGAAACAAACTGCACTTGTTGAGCTTGTGAAACACAAGCCCAAGGCAACAAAAGAGCAACTGAAAGCTGTTATGGATGATTTCGCAGCTTTTGTAGAGAAGTGCTGCAAGGCTGACGATAAGGAGACCTGCTTTGCCGAGGAGGGTAAAAAACTTGTTGCTGCAAGTCGAGCTGCCTTAGGCTTATAA start: 106 end: 1863
Match: ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||
Subjc: GATGCACACAAGAGTGAGGTTGCTCATCGGTTTAAAGATTTGGGAGAAGAAAATTTCAAAGCCTTGGTGTTGATTGCCTTTGCTCAGTATCTTCAGCAGTGTCCATTTGAAGATCATGTAAAATTAGTGAATGAAGTAACTGAATTTGCAAAAACATGTGTTGCTGATGAGTCAGCTGAAAATTGTGACAAATCACTTCATACCCTTTTTGGAGACAAATTATGCACAGTTGCAACTCTTCGTGAAACCTATGGTGAAATGGCTGACTGCTGTGCAAAACAAGAACCTGAGAGAAATGAATGCTTCTTGCAACACAAAGATGACAACCCAAACCTCCCCCGATTGGTGAGACCAGAGGTTGATGTGATGTGCACTGCTTTTCATGACAATGAAGAGACATTTTTGAAAAAATACTTATATGAAATTGCCAGAAGACATCCTTACTTTTATGCCCCGGAACTCCTTTTCTTTGCTAAAAGGTATAAAGCTGCTTTTACAGAATGTTGCCAAGCTGCTGATAAAGCTGCCTGCCTGTTGCCAAAGCTCGATGAACTTCGGGATGAAGGGAAGGCTTCGTCTGCCAAACAGAGACTCAAGTGTGCCAGTCTCCAAAAATTTGGAGAAAGAGCTTTCAAAGCATGGGCAGTAGCTCGCCTGAGCCAGAGATTTCCCAAAGCTGAGTTTGCAGAAGTTTCCAAGTTAGTGACAGATCTTACCAAAGTCCACACGGAATGCTGCCATGGAGATCTGCTTGAATGTGCTGATGACAGGGCGGACCTTGCCAAGTATATCTGTGAAAATCAAGATTCGATCTCCAGTAAACTGAAGGAATGCTGTGAAAAACCTCTGTTGGAAAAATCCCACTGCATTGCCGAAGTGGAAAATGATGAGATGCGTGCTGACTTGCCTTCATTAGCTGCTGATTTTGTTGAAAGTAAGGATGTTTGCAAAAACTATGCTGAGGCAAAGGATGTCTTCCTGGGCATGTTTTTGTATGAATATGCAAGAAGGCATCCTGATTACTCTGTCGTGCTGCTGCTGAGACTTGCCAAGACATATGAAACCACTCTAGAGAAGTGCTGTGCCGCTGCAGATCCTCATGAATGCTATGCCAAAGTGTTCGATGAATTTAAACCTCTTGTGGAAGAGCCTCAGAATTTAATCAAACAAAATTGTGAGCTTTTTGAGCAGCTTGGGGAGTACAAATTCCAGAATGCGCTATTAGTTCGTTACACCAAGAAAGTACCCCAAGTGTCAACTCCAACTCTTGTAGAGGTCTCAAGAAACCTAGGAAAAGTGGGCAGCAAATGTTGTAAACATCCTGAAGCAAAGAGAATGCCCTGTGCAGAAGACTATCTATCCGTGGTCCTGAACCAGTTATGTGTGTTGCATGAGAAAACGCCAGTAAGTGACAGAGTCACCAAATGCTGCACAGAATCCTTGGTGAACAGGCGACCATGCTTTTCAGCTCTGGAAGTCGATGAAACATACGTTCCCAAAGAGTTTAATGCTGAAACATTCACCTTCCATGCAGATATATGCACACTTTCTGAGAAGGAGAGACAAATCAAGAAACAAACTGCACTTGTTGAGCTTGTGAAACACAAGCCCAAGGCAACAAAAGAGCAACTGAAAGCTGTTATGGATGATTTCGCAGCTTTTGTAGAGAAGTGCTGCAAGGCTGACGATAAGGAGACCTGCTTTGCCGAGGAGGGTAAAAAACTTGTTGCTGCAAGTCAAGCTGCCTTAGGCTTATAA start: 1 end: 1758
Align Title:gi|194391111|dbj|AK298461.1| Homo sapiens cDNA FLJ50830 complete cds, highly similar to Serum albumin precursor
Align Len: 1799
Score: 3084.0 Bits: 2782.08 E-val: 0.0
N.aligns:None Ident:1551 Pos.:1551 Gaps:0 Align len:1557
Strand: (None, None) Frame: (1, 1)
Query: AATGAAGAGACATTTTTGAAAAAATACTTATATGAAATTGCCAGAAGACATCCTTACTTCTATGCCCCGGAACTCCTTTTCTTTGCTAAAAGGTATAAAGCTGCTTTTACAGAATGTTGCCAAGCTGCTGATAAGGCTGCCTGCCTGTTGCCAAAGCTCGATGAACTTCGGGATGAAGGGAAGGCTTCGTCTGCCAAACAGAGGCTCAAGTGTGCCAGTCTCCAAAAATTTGGAGAAAGAGCTTTCAAAGCATGGGCAGTAGCTCGCCTGAGCCAGAGATTTCCCAAAGCTGAGTTTGCAGAAGTTTCCAAGTTAGTGACAGATCTTACCAAAGTCCACACGGAATGCTGCCATGGAGATCTGCTTGAATGTGCTGATGACAGGGCGGACCTTGCCAAGTATATCTGTGAAAATCAAGATTCGATCTCCAGTAAACTGAAGGAATGCTGTGAAAAACCTCTGTTGGAAAAATCCCACTGCATTGCCGAAGTGGAAAATGATGAGATGCCTGCTGACTTGCCTTCATTAGCTGCTGATTTTGTTGAAAGTAAGGATGTTTGCAAAAACTATGCTGAGGCAAAGGATGTCTTCCTGGGCATGTTTTTGTATGAATATGCAAGAAGGCATCCTGATTACTCTGTCGTGCTGCTGCTGAGACTTGCCAAGACATATAAAACCACTCTAGAGAAGTGCTGTGCCGCTGCAGATCCTCATGAATGCTATGCCAAAGTGTTCGATGAATTTAAACCTCTTGTGGAAGAGCCTCAGAATTTAATCAAACAAAATTGTGAGCTTTTTGAGCAGCTTGGAGAGTACAAATTCCAGAATGCGCTATTAGTTCGTTACACCAAGAAAGTACCCCAAGTGTCAACTCCAACTCTTGTAGAGGTCTCAAGAAACCTAGGAAAAGTGGGCAGCAAATGTTGTAAACATCCTGAAGCAAAAAGAATGCCCTGTGCAGAAGACTATCTATCCGTGGTCCTGAACCAGTTATGTGTGTTGCATGAGAAAACGCCAGTAAGTGACAGAGTCACCAAATGCTGCACAGAATCCTTGGTGAACAGGCGACCATGCTTTTCAGCTCTGGAAGTCGATGAAACATACGTTCCCAAAGAGTTTAATGCTGAAACATTCACCTTCCATGCAGATATATGCACACTTTCTGAGAAGGAGAGACAAATCAAGAAACAAACTGCACTTGTTGAGCTTGTGAAACACAAGCCCAAGGCAACAAAAGAGCAACTGAAAGCTGTTATGGATGATTTCGCAGCTTTTGTAGAGAAGTGCTGCAAGGCTGACGATAAGGAGACCTGCTTTGCCGAGGAGGGTAAAAAACTTGTTGCTGCAAGTCGAGCTGCCTTAGGCTTATAACATCACATTTAAAAGCATCTCAGCCTACCATGAGAATAAGAGAAAGAAAATGAAGATCAAAAGCTTATTCATCTGTTTTTCTTTTTCGTTGGTGTAAAGCCAACACCCTGTCTAAAAAACATAAATTTCTTTAATCATTTTGCCTCTTTTCTCTGTGCTTCAATTAATAAAAAATGGAAAGAATCT start: 493 end: 2049
Match: ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Subjc: AATGAAGAGACATTTTTGAAAAAATACTTATATGAAATTGCCAGAAGACATCCTTACTTTTATGCCCCGGAACTCCTTTTCTTTGCTAAAAGGTATAAAGCTGCTTTTACAGAATGTTGCCAAGCTGCTGATAAAGCTGCCTGCCTGTTGCCAAAGCTCGATGAACTTCGGGATGAAGGGAAGGCTTCGTCTGCCAAACAGAGACTCAAGTGTGCCAGTCTCCAAAAATTTGGAGAAAGAGCTTTCAAAGCATGGGCAGTAGCTCGCCTGAGCCAGAGATTTCCCAAAGCTGAGTTTGCAGAAGTTTCCAAGTTAGTGACAGATCTTACCAAAGTCCACACGGAATGCTGCCATGGAGATCTGCTTGAATGTGCTGATGACAGGGCGGACCTTGCCAAGTATATCTGTGAAAATCAAGATTCGATCTCCAGTAAACTGAAGGAATGCTGTGAAAAACCTCTGTTGGAAAAATCCCACTGCATTGCCGAAGTGGAAAATGATGGGATGCCTGCTGACTTGCCTTCATTAGCTGCTGATTTTGTTGAAAGTAAGGATGTTTGCAAAAACTATGCTGAGGCAAAGGATGTCTTCCTGGGCATGTTTTTGTATGAATATGCAAGAAGGCATCCTGATTACTCTGTCGTGCTGCTGCTGAGACTTGCCAAGACATATGAAACCACTCTAGAGAAGTGCTGTGCCGCTGCAGATCCTCATGAATGCTATGCCAAAGTGTTCGATGAATTTAAACCTCTTGTGGAAGAGCCTCAGAATTTAATCAAACAAAATTGTGAGCTTTTTGAGCAGCTTGGAGAGTACAAATTCCAGAATGCGCTATTAGTTCGTTACACCAAGAAAGTACCCCAAGTGTCAACTCCAACTCTTGTAGAGGTCTCAAGAAACCTAGGAAAAGTGGGCAGCAAATGTTGTAAACATCCTGAAGCAAAAAGAATGCCCTGTGCAGAAGACTATCTATCCGTGGTCCTGAACCAGTTATGTGTGTTGCATGAGAAAACGCCAGTAAGTGACAGAGTCACCAAATGCTGCACAGAATCCTTGGTGAACAGGCGACCATGCTTTTCAGCTCTGGAAGTCGATGAAACATACGTTCCCAAAGAGTTTAATGCTGAAACATTCACCTTCCATGCAGATATATGCACACTTTCTGAGAAGGAGAGACAAATCAAGAAACAAACTGCACTTGTTGAGCTTGTGAAACACAAGCCCAAGGCAACAAAAGAGCAACTGAAAGCTGTTATGGATGATTTCGCAGCTTTTGTAGAGAAGTGCTGCAAGGCTGACGATAAGGAGACCTGCTTTGCCGAGGAGGGTAAAAAACTTGTTGCTGCAAGTCAAGCTGCCTTAGGCTTATAACATCACATTTAAAAGCATCTCAGCCTACCATGAGAATAAGAGAAAGAAAATGAAGATCAAAAGCTTATTCATCTGTTTTTCTTTTTCGTTGGTGTAAAGCCAACACCCTGTCTAAAAAACATAAATTTCTTTAATCATTTTGCCTCTTTTCTCTGTGCTTCAATTAATAAAAAATGGAAAGAATCT start: 243 end: 1799
Score: 472.0 Bits: 426.881 E-val: 4.90655e-116
N.aligns:None Ident:239 Pos.:239 Gaps:0 Align len:241
Strand: (None, None) Frame: (1, 1)
Query: TCTCTTCTGTCAACCCCACGCGCCTTTGGCACAATGAAGTGGGTAACCTTTATTTCCCTTCTTTTTCTCTTTAGCTCGGCTTATTCCAGGGGTGTGTTTCGTCGAGATGCACACAAGAGTGAGGTTGCTCATCGGTTTAAAGATTTGGGAGAAGAAAATTTCAAAGCCTTAGTGTTGATTGCCTTTGCTCAGTATCTTCAGCAGTGTCCATTTGAAGATCATGTAAAATTAGTGAATGAAG start: 1 end: 241
Match: ||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Subjc: TCTCTTCTGTCAACCCCACACGCCTTTGGCACAATGAAGTGGGTAACCTTTATTTCCCTTCTTTTTCTCTTTAGCTCGGCTTATTCCAGGGGTGTGTTTCGTCGAGATGCACACAAGAGTGAGGTTGCTCATCGGTTTAAAGATTTGGGAGAAGAAAATTTCAAAGCCTTGGTGTTGATTGCCTTTGCTCAGTATCTTCAGCAGTGTCCATTTGAAGATCATGTAAAATTAGTGAATGAAG start: 9 end: 249
Align Title:gi|763428|gb|U22961.1|HSU22961 Human mRNA clone with similarity to L-glycerol-3-phosphate:NAD oxidoreductase and albumin gene sequences
Align Len: 3239
Score: 2748.0 Bits: 2479.12 E-val: 0.0
N.aligns:None Ident:1386 Pos.:1386 Gaps:0 Align len:1394
Strand: (None, None) Frame: (1, 1)
Query: CTGTCAACCCCACGCGCCTTTGGCACAATGAAGTGGGTAACCTTTATTTCCCTTCTTTTTCTCTTTAGCTCGGCTTATTCCAGGGGTGTGTTTCGTCGAGATGCACACAAGAGTGAGGTTGCTCATCGGTTTAAAGATTTGGGAGAAGAAAATTTCAAAGCCTTAGTGTTGATTGCCTTTGCTCAGTATCTTCAGCAGTGTCCATTTGAAGATCATGTAAAATTAGTGAATGAAGTAACTGAATTTGCAAAAACATGTGTTGCTGATGAGTCAGCTGAAAATTGTGACAAATCACTTCATACCCTTTTTGGAGACAAATTATGCACAGTTGCAACTCTTCGTGAAACCTATGGTGAAATGGCTGACTGCTGTGCAAAACAAGAACCTGAGAGAAATGAATGCTTCTTGCAACACAAAGATGACAACCCAAACCTCCCCCGATTGGTGAGACCAGAGGTTGATGTGATGTGCACTGCTTTTCATGACAATGAAGAGACATTTTTGAAAAAATACTTATATGAAATTGCCAGAAGACATCCTTACTTCTATGCCCCGGAACTCCTTTTCTTTGCTAAAAGGTATAAAGCTGCTTTTACAGAATGTTGCCAAGCTGCTGATAAGGCTGCCTGCCTGTTGCCAAAGCTCGATGAACTTCGGGATGAAGGGAAGGCTTCGTCTGCCAAACAGAGGCTCAAGTGTGCCAGTCTCCAAAAATTTGGAGAAAGAGCTTTCAAAGCATGGGCAGTAGCTCGCCTGAGCCAGAGATTTCCCAAAGCTGAGTTTGCAGAAGTTTCCAAGTTAGTGACAGATCTTACCAAAGTCCACACGGAATGCTGCCATGGAGATCTGCTTGAATGTGCTGATGACAGGGCGGACCTTGCCAAGTATATCTGTGAAAATCAAGATTCGATCTCCAGTAAACTGAAGGAATGCTGTGAAAAACCTCTGTTGGAAAAATCCCACTGCATTGCCGAAGTGGAAAATGATGAGATGCCTGCTGACTTGCCTTCATTAGCTGCTGATTTTGTTGAAAGTAAGGATGTTTGCAAAAACTATGCTGAGGCAAAGGATGTCTTCCTGGGCATGTTTTTGTATGAATATGCAAGAAGGCATCCTGATTACTCTGTCGTGCTGCTGCTGAGACTTGCCAAGACATATAAAACCACTCTAGAGAAGTGCTGTGCCGCTGCAGATCCTCATGAATGCTATGCCAAAGTGTTCGATGAATTTAAACCTCTTGTGGAAGAGCCTCAGAATTTAATCAAACAAAATTGTGAGCTTTTTGAGCAGCTTGGAGAGTACAAATTCCAGAATGCGCTATTAGTTCGTTACACCAAGAAAGTACCCCAAGTGTCAACTCCAACTCTTGTAGAGGTCTCAAGAAACCTAGGAAA start: 7 end: 1400
Match: ||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Subjc: CTGTCAACCCCACACGCCTTTGGCACAATGAAGTGGGTAACCTTTATTTCCCTTCTTTTTCTCTTTAGCTCGGCTTATTCCAGGGGTGTGTTTCGTCGAGATGCACACAAGAGTGAGGTTGCTCATCGGTTTAAAGATTTGGGAGAAGAAAATTTCAAAGCCTTGGTGTTGATTGCCTTTGCTCAGTATCTTCAGCAGTGTCCATTTGAAGATCATGTAAAATTAGTGAATGAAGTAACTGAATTTGCAAAAACATGTGTTGCTGATGAGTCAGCTGAAAATTGTGACAAATCACTTCATACCCTTTTTGGAGACAAATTATGCACAGTTGCAACTCTTCGTGAAACCTATGGTGAAATGGCTGACTGCTGTGCAAAACAAGAACCTGGGAGAAATGAATGCTTCTTGCAACACAAAGATGACAACCCAAACCTCCCCCGATTGGTGAGACCAGAGGTTGATGTGATGTGCACTGCTTTTCATGACAATGAAGAGACATTTTTGAAAAAATACTTATATGAAATTGCCAGAAGACATCCTTACTTTTATGCCCCGGAACTCCTTTTCTTTGCTAAAAGGTATAAAGCTGCTTTTACAGAATGTTGCCAAGCTGCTGATAAAGCTGCCTGCCTGTTGCCAAAGCTCGATGAACTTCGGGATGAAGGGAAGGCTTCGTCTGCCAAACAGAGACTCAAGTGTGCCAGTCTCCAAAAATTTGGAGAAAGAGCTTTCAAAGCATGGGCAGTAGCTCGCCTGAGCCAGAGATTTCCCAAAGCTGAGTTTGCAGAAGTTTCCAAGTTAGTGACAGATCTTACCAAAGTCCACACGGAATGCTGCCATGGAGATCTGCTTGAATGTGCTGATGACAGGGCGGACCTTGCCAAGTATATCTGTGAAAATCAAGATTCGATCTCCAGTAAACTGAAGGAATGCTGTGAAAAACCTCTGTTGGAAAAATCCCACTGCATTGCCGAAGTGGAAAATGATGAGATGCCTGCTGACTTGCCTTCATTAGCTGCTGATTTTGTTGAAAGTAAGGATGTTTGCAAAAACTATGCTGAGGCAAAGGATGTCTTCTTGGGCATGTTTTTGTATGAATATGCAAGAAGGCATCCTGATTACTCTGTCGTGCTGCTGCTGAGACTTGCCAAGACATATGAAACCACTCTAGAGAAGTGCTGTGCCGCTGCAGATCCTCATGAATGCTATGCCAAAGTGTTCGATGAATTTAAACCTCTTGTGGAAGAGCCTCAGAATTTAATCAAACAAAATTGTGAGCTTTTTGAGCAGCTTGGAGAGTACAAATTCCAGAATGCGCTATTAGTTCGTTACACCAAGAAAGTACCCCAAGTGTCAACTCCAACTCTTGTAGAGGTCTCAAGAAACCTAGGAAA start: 1846 end: 3239
Align Title:gi|27692692|gb|BC041789.1| Homo sapiens albumin, mRNA (cDNA clone MGC:32888 IMAGE:4766983), complete cds
Align Len: 1510
Score: 2620.0 Bits: 2363.7 E-val: 0.0
N.aligns:None Ident:1327 Pos.:1327 Gaps:1 Align len:1337
Strand: (None, None) Frame: (1, 1)
Query: AAAAATTTGG-AGAAAGAGCTTTCAAAGCATGGGCAGTAGCTCGCCTGAGCCAGAGATTTCCCAAAGCTGAGTTTGCAGAAGTTTCCAAGTTAGTGACAGATCTTACCAAAGTCCACACGGAATGCTGCCATGGAGATCTGCTTGAATGTGCTGATGACAGGGCGGACCTTGCCAAGTATATCTGTGAAAATCAAGATTCGATCTCCAGTAAACTGAAGGAATGCTGTGAAAAACCTCTGTTGGAAAAATCCCACTGCATTGCCGAAGTGGAAAATGATGAGATGCCTGCTGACTTGCCTTCATTAGCTGCTGATTTTGTTGAAAGTAAGGATGTTTGCAAAAACTATGCTGAGGCAAAGGATGTCTTCCTGGGCATGTTTTTGTATGAATATGCAAGAAGGCATCCTGATTACTCTGTCGTGCTGCTGCTGAGACTTGCCAAGACATATAAAACCACTCTAGAGAAGTGCTGTGCCGCTGCAGATCCTCATGAATGCTATGCCAAAGTGTTCGATGAATTTAAACCTCTTGTGGAAGAGCCTCAGAATTTAATCAAACAAAATTGTGAGCTTTTTGAGCAGCTTGGAGAGTACAAATTCCAGAATGCGCTATTAGTTCGTTACACCAAGAAAGTACCCCAAGTGTCAACTCCAACTCTTGTAGAGGTCTCAAGAAACCTAGGAAAAGTGGGCAGCAAATGTTGTAAACATCCTGAAGCAAAAAGAATGCCCTGTGCAGAAGACTATCTATCCGTGGTCCTGAACCAGTTATGTGTGTTGCATGAGAAAACGCCAGTAAGTGACAGAGTCACCAAATGCTGCACAGAATCCTTGGTGAACAGGCGACCATGCTTTTCAGCTCTGGAAGTCGATGAAACATACGTTCCCAAAGAGTTTAATGCTGAAACATTCACCTTCCATGCAGATATATGCACACTTTCTGAGAAGGAGAGACAAATCAAGAAACAAACTGCACTTGTTGAGCTTGTGAAACACAAGCCCAAGGCAACAAAAGAGCAACTGAAAGCTGTTATGGATGATTTCGCAGCTTTTGTAGAGAAGTGCTGCAAGGCTGACGATAAGGAGACCTGCTTTGCCGAGGAGGGTAAAAAACTTGTTGCTGCAAGTCGAGCTGCCTTAGGCTTATAACATCACATTTAAAAGCATCTCAGCCTACCATGAGAATAAGAGAAAGAAAATGAAGATCAAAAGCTTATTCATCTGTTTTTCTTTTTCGTTGGTGTAAAGCCAACACCCTGTCTAAAAAACATAAATTTCTTTAATCATTTTGCCTCTTTTCTCTGTGCTTCAATTAATAAAAAATGGAAAGAATCTAA start: 716 end: 2051
Match: ||| |||||| |||| | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| ||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| ||
Subjc: AAAGATTTGGGAGAAGAAAATTTCAAAGCATGGGCAGTAGCTCGCCTGAGCCAGAGATTTCCCAAAGCTGAGTTTGCAGAAGTTTCCAAGTTAGTGACAGATCTTACCAAAGTCCACACGGAATGCTGCCATGGAGATCTGCTTGAATGTGCTGATGACAGGGCGGACCTTGCCAAGTATATCTGTGAAAATCAAGATTCGATCTCCAGTAAACTGAAGGAATGCTGTGAAAAACCTCTGTTGGAAAAATCCCACTGCATTGCCGAAGTGGAAAATGATGAGATGCCTGCTGACTTGCCTTCATTAGCTGCTGATTTTGTTGAAAGTAAGGATGTTTGCAAAAACTATGCTGAGGCAAAGGATGTCTTCCTGGGCATGTTTTTGTATGAATATGCAAGAAGGCATCCTGATTACTCTGTCGTGCTGCTGCTGAGACTTGCCAAGACATATGAAACCACTCTAGAGAAGTGCTGTGCCGCTGCGGATCCTCATGAATGCTATGCCAAAGTGTTCGATGAATTTAAACCTCTTGTGGAAGAGCCTCAGAATTTAATCAAACAAAATTGTGAGCTTTTTGAGCAGCTTGGAGAGTACAAATTCCAGAATGCGCTATTAGTTCGTTACACCAAGAAAGTACCCCAAGTGTCAACTCCAACTCTTGTAGAGGTCTCAAGAAACCTAGGAAAAGTGGGCAGCAAATGTTGTAAACATCCTGAAGCAAAAAGAATGCCCTGTGCAGAAGACTATCTATCCGTGGTCCTGAACCAGTTATGTGTGTTGCATGAGAAAACGCCAGTAAGTGACAGAGTCACCAAATGCTGCACAGAATCCTTGGTGAACAGGCGACCATGCTTTTCAGCTCTGGAAGTCGATGAAACATACGTTCCCAAAGAGTTTAATGCTGAAACATTCACCTTCCATGCAGATATATGCACACTTTCTGAGAAGGAGAGACAAATCAAGAAACAAACTGCACTTGTTGAGCTTGTGAAACACAAGCCCAAGGCAACAAAAGAGCAACTGAAAGCTGTTATGGATGATTTCGCAGCTTTTGTAGAGAAGTGCTGCAAGGCTGACGATAAGGAGACCTGCTTTGCCGAGGAGGGTAAAAAACTTGTTGCTGCAAGTCAAGCTGCCTTAGGCTTATAACATCACATTTAAAAGCATCTCAGCCTACCATGAGAATAAGAGAAAGAAAATGAAGATCAAAAGCTTATTCATCTGTTTTTCTTTTTCGTTGGTGTAAAGCCAACACCCTGTCTAAAAAACATAAATTTCTTTAATCATTTTGCCTCTTTTCTCTGTGCTTCAATTAATAAAAAATGGAAAGAATCGAA start: 147 end: 1483
Score: 329.0 Bits: 297.94 E-val: 5.98796e-77
N.aligns:None Ident:166 Pos.:166 Gaps:0 Align len:167
Strand: (None, None) Frame: (1, 1)
Query: TCTCTTCTGTCAACCCCACGCGCCTTTGGCACAATGAAGTGGGTAACCTTTATTTCCCTTCTTTTTCTCTTTAGCTCGGCTTATTCCAGGGGTGTGTTTCGTCGAGATGCACACAAGAGTGAGGTTGCTCATCGGTTTAAAGATTTGGGAGAAGAAAATTTCAAAGC start: 1 end: 167
Match: ||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Subjc: TCTCTTCTGTCAACCCCACACGCCTTTGGCACAATGAAGTGGGTAACCTTTATTTCCCTTCTTTTTCTCTTTAGCTCGGCTTATTCCAGGGGTGTGTTTCGTCGAGATGCACACAAGAGTGAGGTTGCTCATCGGTTTAAAGATTTGGGAGAAGAAAATTTCAAAGC start: 9 end: 175
Align Title:gi|164694201|dbj|AK308044.1| Homo sapiens cDNA, FLJ97992
Align Len: 1544
Score: 2536.0 Bits: 2287.96 E-val: 0.0
N.aligns:None Ident:1279 Pos.:1279 Gaps:1 Align len:1285
Strand: (None, None) Frame: (1, 1)
Query: TGCAAAACAAGAACCTGAGAGAAATGAATGCTTCTTGCAACACAAAGATGACAACCCAAACCTCCCCCGATTGGTGAGACCAGAGGTTGATGTGATGTGCACTGCTTTTCATGACAATGAAGAGACATTTTTGAAAAAATACTTATATGAAATTGCCAGAAGACATCCTTACTTCTATGCCCCGGAACTCCTTTTCTTTGCTAAAAGGTATAAAGCTGCTTTTACAGAATGTTGCCAAGCTGCTGATAAGGCTGCCTGCCTGTTGCCAAAGCTCGATGAACTTCGGGATGAAGGGAAGGCTTCGTCTGCCAAACAGAGGCTCAAGTGTGCCAGTCTCCAAAAATTTGGAGAAAGAGCTTTCAAAGCATGGGCAGTAGCTCGCCTGAGCCAGAGATTTCCCAAAGCTGAGTTTGCAGAAGTTTCCAAGTTAGTGACAGATCTTACC-AAAGTCCACACGGAATGCTGCCATGGAGATCTGCTTGAATGTGCTGATGACAGGGCGGACCTTGCCAAGTATATCTGTGAAAATCAAGATTCGATCTCCAGTAAACTGAAGGAATGCTGTGAAAAACCTCTGTTGGAAAAATCCCACTGCATTGCCGAAGTGGAAAATGATGAGATGCCTGCTGACTTGCCTTCATTAGCTGCTGATTTTGTTGAAAGTAAGGATGTTTGCAAAAACTATGCTGAGGCAAAGGATGTCTTCCTGGGCATGTTTTTGTATGAATATGCAAGAAGGCATCCTGATTACTCTGTCGTGCTGCTGCTGAGACTTGCCAAGACATATAAAACCACTCTAGAGAAGTGCTGTGCCGCTGCAGATCCTCATGAATGCTATGCCAAAGTGTTCGATGAATTTAAACCTCTTGTGGAAGAGCCTCAGAATTTAATCAAACAAAATTGTGAGCTTTTTGAGCAGCTTGGAGAGTACAAATTCCAGAATGCGCTATTAGTTCGTTACACCAAGAAAGTACCCCAAGTGTCAACTCCAACTCTTGTAGAGGTCTCAAGAAACCTAGGAAAAGTGGGCAGCAAATGTTGTAAACATCCTGAAGCAAAAAGAATGCCCTGTGCAGAAGACTATCTATCCGTGGTCCTGAACCAGTTATGTGTGTTGCATGAGAAAACGCCAGTAAGTGACAGAGTCACCAAATGCTGCACAGAATCCTTGGTGAACAGGCGACCATGCTTTTCAGCTCTGGAAGTCGATGAAACATACGTTCCCAAAGAGTTTAATGCTGAAACATTCACCTTCCATGCAGATATATGCACACTTTCTGAGAA start: 378 end: 1661
Match: |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| || |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Subjc: TGCAAAACAAGAACCTGAGAGAAATGAATGCTTCTTGCAACACAAAGATGACAACCCAAACCTCCCCCGATTGGTGAGACCAGAGGTTGATGTGATGTGCACTGCTTTTCATGACAATGAAGAGACATTTTTGAAAAAATACTTATATGAAATTGCCAGAAGACATCCTTACTTTTATGCCCCGGAACTCCTTTTCTTTGCTAAAAGGTATAAAGCTGCTTTTACAGAATGTTGCCAAGCTGCTGATAAAGCTGCCTGCCTGTTGCCAAAGCTCGATGAACTTCGGGATGAAGGGAAGGCTTCGTCTGCCAAACAGAGACTCAAGTGTGCCAGTCTCCAAAAATTTGGAGAAAGAGCTTTCAAAGCATGGGCAGTAGCTCGCCTGAGCCAGAGATTTCCCAAAGCTGAGTTTGCAGAAGTTTCCAAGTTAGTGACAGATCTTTCCCAAAGTCCACACGGAATGCTGCCATGGAGATCTGCTTGAATGTGCTGATGACAGGGCGGACCTTGCCAAGTATATCTGTGAAAATCAAGATTCGATCTCCAGTAAACTGAAGGAATGCTGTGAAAAACCTCTGTTGGAAAAATCCCACTGCATTGCCGAAGTGGAAAATGATGAGATGCCTGCTGACTTGCCTTCATTAGCTGCTGATTTTGTTGAAAGTAAGGATGTTTGCAAAAACTATGCTGAGGCAAAGGATGTCTTCCTGGGCATGTTTTTGTATGAATATGCAAGAAGGCATCCTGATTACTCTGTCGTGCTGCTGCTGAGACTTGCCAAGACATATGAAACCACTCTAGAGAAGTGCTGTGCCGCTGCAGATCCTCATGAATGCTATGCCAAAGTGTTCGATGAATTTAAACCTCTTGTGGAAGAGCCTCAGAATTTAATCAAACAAAATTGTGAGCTTTTTGAGCAGCTTGGAGAGTACAAATTCCAGAATGCGCTATTAGTTCGTTACACCAAGAAAGTACCCCAAGTGTCAACTCCAACTCTTGTAGAGGTCTCAAGAAACCTAGGAAAAGTGGGCAGCAAATGTTGTAAACATCCTGAAGCAAAAAGAATGCCCTGTGCAGAAGACTATCTATCCGTGGTCCTGAACCAGTTATGTGTGTTGCATGAGAAAACGCCAGTAAGTGACAGAGTCACCAAATGCTGCACAGAATCCTTGGTGAACAGGCGACCATGCTTTTCAGCTCTGGAAGTCGATGAAACATACGTTCCCAAAGAGTTTAATGCTGAAACATTCACCTTCCATGCAGATATATGCACACTTTCTGAGAA start: 260 end: 1544
Score: 506.0 Bits: 457.539 E-val: 2.89746e-125
N.aligns:None Ident:256 Pos.:256 Gaps:0 Align len:258
Strand: (None, None) Frame: (1, 1)
Query: TCTCTTCTGTCAACCCCACGCGCCTTTGGCACAATGAAGTGGGTAACCTTTATTTCCCTTCTTTTTCTCTTTAGCTCGGCTTATTCCAGGGGTGTGTTTCGTCGAGATGCACACAAGAGTGAGGTTGCTCATCGGTTTAAAGATTTGGGAGAAGAAAATTTCAAAGCCTTAGTGTTGATTGCCTTTGCTCAGTATCTTCAGCAGTGTCCATTTGAAGATCATGTAAAATTAGTGAATGAAGTAACTGAATTTGCAAAA start: 1 end: 258
Match: ||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Subjc: TCTCTTCTGTCAACCCCACACGCCTTTGGCACAATGAAGTGGGTAACCTTTATTTCCCTTCTTTTTCTCTTTAGCTCGGCTTATTCCAGGGGTGTGTTTCGTCGAGATGCACACAAGAGTGAGGTTGCTCATCGGTTTAAAGATTTGGGAGAAGAAAATTTCAAAGCCTTGGTGTTGATTGCCTTTGCTCAGTATCTTCAGCAGTGTCCATTTGAAGATCATGTAAAATTAGTGAATGAAGTAACTGAATTTGCAAAA start: 9 end: 266
Align Title:gi|7770216|gb|AF119890.1|AF119890 Homo sapiens PRO2675 mRNA, complete cds
Align Len: 1618
Score: 2488.0 Bits: 2244.68 E-val: 0.0
N.aligns:None Ident:1247 Pos.:1247 Gaps:0 Align len:1249
Strand: (None, None) Frame: (1, 1)
Query: AGTTAGTGACAGATCTTACCAAAGTCCACACGGAATGCTGCCATGGAGATCTGCTTGAATGTGCTGATGACAGGGCGGACCTTGCCAAGTATATCTGTGAAAATCAAGATTCGATCTCCAGTAAACTGAAGGAATGCTGTGAAAAACCTCTGTTGGAAAAATCCCACTGCATTGCCGAAGTGGAAAATGATGAGATGCCTGCTGACTTGCCTTCATTAGCTGCTGATTTTGTTGAAAGTAAGGATGTTTGCAAAAACTATGCTGAGGCAAAGGATGTCTTCCTGGGCATGTTTTTGTATGAATATGCAAGAAGGCATCCTGATTACTCTGTCGTGCTGCTGCTGAGACTTGCCAAGACATATAAAACCACTCTAGAGAAGTGCTGTGCCGCTGCAGATCCTCATGAATGCTATGCCAAAGTGTTCGATGAATTTAAACCTCTTGTGGAAGAGCCTCAGAATTTAATCAAACAAAATTGTGAGCTTTTTGAGCAGCTTGGAGAGTACAAATTCCAGAATGCGCTATTAGTTCGTTACACCAAGAAAGTACCCCAAGTGTCAACTCCAACTCTTGTAGAGGTCTCAAGAAACCTAGGAAAAGTGGGCAGCAAATGTTGTAAACATCCTGAAGCAAAAAGAATGCCCTGTGCAGAAGACTATCTATCCGTGGTCCTGAACCAGTTATGTGTGTTGCATGAGAAAACGCCAGTAAGTGACAGAGTCACCAAATGCTGCACAGAATCCTTGGTGAACAGGCGACCATGCTTTTCAGCTCTGGAAGTCGATGAAACATACGTTCCCAAAGAGTTTAATGCTGAAACATTCACCTTCCATGCAGATATATGCACACTTTCTGAGAAGGAGAGACAAATCAAGAAACAAACTGCACTTGTTGAGCTTGTGAAACACAAGCCCAAGGCAACAAAAGAGCAACTGAAAGCTGTTATGGATGATTTCGCAGCTTTTGTAGAGAAGTGCTGCAAGGCTGACGATAAGGAGACCTGCTTTGCCGAGGAGGGTAAAAAACTTGTTGCTGCAAGTCGAGCTGCCTTAGGCTTATAACATCACATTTAAAAGCATCTCAGCCTACCATGAGAATAAGAGAAAGAAAATGAAGATCAAAAGCTTATTCATCTGTTTTTCTTTTTCGTTGGTGTAAAGCCAACACCCTGTCTAAAAAACATAAATTTCTTTAATCATTTTGCCTCTTTTCTCTGTGCTTCAATTAATAAAAAATGGAAAGAATCTAA start: 803 end: 2051
Match: |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Subjc: AGTTAGTGACAGATCTTACCAAAGTCCACACGGAATGCTGCCATGGAGATCTGCTTGAATGTGCTGATGACAGGGCGGACCTTGCCAAGTATATCTGTGAAAATCAAGATTCGATCTCCAGTAAACTGAAGGAATGCTGTGAAAAACCTCTGTTGGAAAAATCCCACTGCATTGCCGAAGTGGAAAATGATGAGATGCCTGCTGACTTGCCTTCATTAGCTGCTGATTTTGTTGAAAGTAAGGATGTTTGCAAAAACTATGCTGAGGCAAAGGATGTCTTCCTGGGCATGTTTTTGTATGAATATGCAAGAAGGCATCCTGATTACTCTGTCGTGCTGCTGCTGAGACTTGCCAAGACATATGAAACCACTCTAGAGAAGTGCTGTGCCGCTGCAGATCCTCATGAATGCTATGCCAAAGTGTTCGATGAATTTAAACCTCTTGTGGAAGAGCCTCAGAATTTAATCAAACAAAATTGTGAGCTTTTTGAGCAGCTTGGAGAGTACAAATTCCAGAATGCGCTATTAGTTCGTTACACCAAGAAAGTACCCCAAGTGTCAACTCCAACTCTTGTAGAGGTCTCAAGAAACCTAGGAAAAGTGGGCAGCAAATGTTGTAAACATCCTGAAGCAAAAAGAATGCCCTGTGCAGAAGACTATCTATCCGTGGTCCTGAACCAGTTATGTGTGTTGCATGAGAAAACGCCAGTAAGTGACAGAGTCACCAAATGCTGCACAGAATCCTTGGTGAACAGGCGACCATGCTTTTCAGCTCTGGAAGTCGATGAAACATACGTTCCCAAAGAGTTTAATGCTGAAACATTCACCTTCCATGCAGATATATGCACACTTTCTGAGAAGGAGAGACAAATCAAGAAACAAACTGCACTTGTTGAGCTTGTGAAACACAAGCCCAAGGCAACAAAAGAGCAACTGAAAGCTGTTATGGATGATTTCGCAGCTTTTGTAGAGAAGTGCTGCAAGGCTGACGATAAGGAGACCTGCTTTGCCGAGGAGGGTAAAAAACTTGTTGCTGCAAGTCAAGCTGCCTTAGGCTTATAACATCACATTTAAAAGCATCTCAGCCTACCATGAGAATAAGAGAAAGAAAATGAAGATCAAAAGCTTATTCATCTGTTTTTCTTTTTCGTTGGTGTAAAGCCAACACCCTGTCTAAAAAACATAAATTTCTTTAATCATTTTGCCTCTTTTCTCTGTGCTTCAATTAATAAAAAATGGAAAGAATCTAA start: 350 end: 1598
Align Title:gi|47118014|gb|AY544124.1| Homo sapiens growth-inhibiting protein 20 mRNA, complete cds
Align Len: 1363
Score: 2453.0 Bits: 2213.12 E-val: 0.0
N.aligns:None Ident:1242 Pos.:1242 Gaps:1 Align len:1251
Strand: (None, None) Frame: (1, 1)
Query: AAAAATTTGG-AGAAAGAGCTTTCAAAGCATGGGCAGTAGCTCGCCTGAGCCAGAGATTTCCCAAAGCTGAGTTTGCAGAAGTTTCCAAGTTAGTGACAGATCTTACCAAAGTCCACACGGAATGCTGCCATGGAGATCTGCTTGAATGTGCTGATGACAGGGCGGACCTTGCCAAGTATATCTGTGAAAATCAAGATTCGATCTCCAGTAAACTGAAGGAATGCTGTGAAAAACCTCTGTTGGAAAAATCCCACTGCATTGCCGAAGTGGAAAATGATGAGATGCCTGCTGACTTGCCTTCATTAGCTGCTGATTTTGTTGAAAGTAAGGATGTTTGCAAAAACTATGCTGAGGCAAAGGATGTCTTCCTGGGCATGTTTTTGTATGAATATGCAAGAAGGCATCCTGATTACTCTGTCGTGCTGCTGCTGAGACTTGCCAAGACATATAAAACCACTCTAGAGAAGTGCTGTGCCGCTGCAGATCCTCATGAATGCTATGCCAAAGTGTTCGATGAATTTAAACCTCTTGTGGAAGAGCCTCAGAATTTAATCAAACAAAATTGTGAGCTTTTTGAGCAGCTTGGAGAGTACAAATTCCAGAATGCGCTATTAGTTCGTTACACCAAGAAAGTACCCCAAGTGTCAACTCCAACTCTTGTAGAGGTCTCAAGAAACCTAGGAAAAGTGGGCAGCAAATGTTGTAAACATCCTGAAGCAAAAAGAATGCCCTGTGCAGAAGACTATCTATCCGTGGTCCTGAACCAGTTATGTGTGTTGCATGAGAAAACGCCAGTAAGTGACAGAGTCACCAAATGCTGCACAGAATCCTTGGTGAACAGGCGACCATGCTTTTCAGCTCTGGAAGTCGATGAAACATACGTTCCCAAAGAGTTTAATGCTGAAACATTCACCTTCCATGCAGATATATGCACACTTTCTGAGAAGGAGAGACAAATCAAGAAACAAACTGCACTTGTTGAGCTTGTGAAACACAAGCCCAAGGCAACAAAAGAGCAACTGAAAGCTGTTATGGATGATTTCGCAGCTTTTGTAGAGAAGTGCTGCAAGGCTGACGATAAGGAGACCTGCTTTGCCGAGGAGGGTAAAAAACTTGTTGCTGCAAGTCGAGCTGCCTTAGGCTTATAACATCACATTTAAAAGCATCTCAGCCTACCATGAGAATAAGAGAAAGAAAATGAAGATCAAAAGCTTATTCATCTGTTTTTCTTTTTCGTTGGTGTAAAGCCA start: 716 end: 1965
Match: ||| |||||| |||| | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| ||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Subjc: AAAGATTTGGGAGAAGAAAATTTCAAAGCATGGGCAGTAGCTCGCCTGAGCCAGAGATTTCCCAAAGCTGAGTTTGCAGAAGTTTCCAAGTTAGTGACAGATCTTACCAAAGTCCACACGGAATGCTGCCATGGAGATCTGCTTGAATGTGCTGATGACAGGGCGGACCTTGCCAAGTATATCTGTGAAAATCAAGATTCGATCTCCAGTAAACTGAAGGAATGCTGTGAAAAACCTCTGTTGGAAAAATCCCACTGCATTGCCGAAGTGGAAAATGATGAGATGCCTGCTGACTTGCCTTCATTAGCTGCTGATTTTGTTGAAAGTAAGGATGTTTGCAAAAACTATGCTGAGGCAAAGGATGTCTTCCTGGGCATGTTTTTGTATGAATATGCAAGAAGGCATCCTGATTACTCTGTCGTGCTGCTGCTGAGACTTGCCAAGACATATGAAACCACTCTAGAGAAGTGCTGTGCCGCTGCGGATCCTCATGAATGCTATGCCAAAGTGTTCGATGAATTTAAACCTCTTGTGGAAGAGCCTCAGAATTTAATCAAACAAAATTGTGAGCTTTTTGAGCAGCTTGGAGAGTACAAATTCCAGAATGCGCTATTAGTTCGTTACACCAAGAAAGTACCCCAAGTGTCAACTCCAACTCTTGTAGAGGTCTCAAGAAACCTAGGAAAAGTGGGCAGCAAATGTTGTAAACATCCTGAAGCAAAAAGAATGCCCTGTGCAGAAGACTATCTATCCGTGGTCCTGAACCAGTTATGTGTGTTGCATGAGAAAACGCCAGTAAGTGACAGAGTCACCAAATGCTGCACAGAATCCTTGGTGAACAGGCGACCATGCTTTTCAGCTCTGGAAGTCGATGAAACATACGTTCCCAAAGAGTTTAATGCTGAAACATTCACCTTCCATGCAGATATATGCACACTTTCTGAGAAGGAGAGACAAATCAAGAAACAAACTGCACTTGTTGAGCTTGTGAAACACAAGCCCAAGGCAACAAAAGAGCAACTGAAAGCTGTTATGGATGATTTCGCAGCTTTTGTAGAGAAGTGCTGCAAGGCTGACGATAAGGAGACCTGCTTTGCCGAGGAGGGTAAAAAACTTGTTGCTGCAAGTCAAGCTGCCTTAGGCTTATAACATCACATTTAAAAGCATCTCAGCCTACCATGAGAATAAGAGAAAGAAAATGAAGATCAAAAGCTTATTCATCTGTTTTTCTTTTTCGTTGGTGTAAAGCCA start: 113 end: 1363
Score: 282.0 Bits: 255.561 E-val: 1.83335e-64
N.aligns:None Ident:141 Pos.:141 Gaps:0 Align len:141
Strand: (None, None) Frame: (1, 1)
Query: TGGCACAATGAAGTGGGTAACCTTTATTTCCCTTCTTTTTCTCTTTAGCTCGGCTTATTCCAGGGGTGTGTTTCGTCGAGATGCACACAAGAGTGAGGTTGCTCATCGGTTTAAAGATTTGGGAGAAGAAAATTTCAAAGC start: 27 end: 167
Match: |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Subjc: TGGCACAATGAAGTGGGTAACCTTTATTTCCCTTCTTTTTCTCTTTAGCTCGGCTTATTCCAGGGGTGTGTTTCGTCGAGATGCACACAAGAGTGAGGTTGCTCATCGGTTTAAAGATTTGGGAGAAGAAAATTTCAAAGC start: 1 end: 141
Align Title:gi|7959790|gb|AF116645.1|AF116645 Homo sapiens PRO1708 mRNA, complete cds
Align Len: 1251
Score: 2003.0 Bits: 1807.36 E-val: 0.0
N.aligns:None Ident:1006 Pos.:1006 Gaps:0 Align len:1009
Strand: (None, None) Frame: (1, 1)
Query: TTTTGTTGAAAGTAAGGATGTTTGCAAAAACTATGCTGAGGCAAAGGATGTCTTCCTGGGCATGTTTTTGTATGAATATGCAAGAAGGCATCCTGATTACTCTGTCGTGCTGCTGCTGAGACTTGCCAAGACATATAAAACCACTCTAGAGAAGTGCTGTGCCGCTGCAGATCCTCATGAATGCTATGCCAAAGTGTTCGATGAATTTAAACCTCTTGTGGAAGAGCCTCAGAATTTAATCAAACAAAATTGTGAGCTTTTTGAGCAGCTTGGAGAGTACAAATTCCAGAATGCGCTATTAGTTCGTTACACCAAGAAAGTACCCCAAGTGTCAACTCCAACTCTTGTAGAGGTCTCAAGAAACCTAGGAAAAGTGGGCAGCAAATGTTGTAAACATCCTGAAGCAAAAAGAATGCCCTGTGCAGAAGACTATCTATCCGTGGTCCTGAACCAGTTATGTGTGTTGCATGAGAAAACGCCAGTAAGTGACAGAGTCACCAAATGCTGCACAGAATCCTTGGTGAACAGGCGACCATGCTTTTCAGCTCTGGAAGTCGATGAAACATACGTTCCCAAAGAGTTTAATGCTGAAACATTCACCTTCCATGCAGATATATGCACACTTTCTGAGAAGGAGAGACAAATCAAGAAACAAACTGCACTTGTTGAGCTTGTGAAACACAAGCCCAAGGCAACAAAAGAGCAACTGAAAGCTGTTATGGATGATTTCGCAGCTTTTGTAGAGAAGTGCTGCAAGGCTGACGATAAGGAGACCTGCTTTGCCGAGGAGGGTAAAAAACTTGTTGCTGCAAGTCGAGCTGCCTTAGGCTTATAACATCACATTTAAAAGCATCTCAGCCTACCATGAGAATAAGAGAAAGAAAATGAAGATCAAAAGCTTATTCATCTGTTTTTCTTTTTCGTTGGTGTAAAGCCAACACCCTGTCTAAAAAACATAAATTTCTTTAATCATTTTGCCTCTTTTCTCTGTGCTTCAATTAATAAAAAA start: 1029 end: 2037
Match: |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Subjc: TTTTGTTGAAAGTAAGGATGTTTGCAAAAACTATGCTGAGGCAAAGGATGTCTTCCTGGGCATGTTTTTGTATGAATATGCAAGAAGGCATCCTGATTACTCTGTCGTGCTGCTGCTGAGACTTGCCAAGACATATGAAACCACTCTAGAGAAGTGCTGTGCCGCTGCAGATCCTCATGAATGCTATGCCAAAGTGTTCGATGAATTTAAACCTCTTGCGGAAGAGCCTCAGAATTTAATCAAACAAAATTGTGAGCTTTTTGAGCAGCTTGGAGAGTACAAATTCCAGAATGCGCTATTAGTTCGTTACACCAAGAAAGTACCCCAAGTGTCAACTCCAACTCTTGTAGAGGTCTCAAGAAACCTAGGAAAAGTGGGCAGCAAATGTTGTAAACATCCTGAAGCAAAAAGAATGCCCTGTGCAGAAGACTATCTATCCGTGGTCCTGAACCAGTTATGTGTGTTGCATGAGAAAACGCCAGTAAGTGACAGAGTCACCAAATGCTGCACAGAATCCTTGGTGAACAGGCGACCATGCTTTTCAGCTCTGGAAGTCGATGAAACATACGTTCCCAAAGAGTTTAATGCTGAAACATTCACCTTCCATGCAGATATATGCACACTTTCTGAGAAGGAGAGACAAATCAAGAAACAAACTGCACTTGTTGAGCTTGTGAAACACAAGCCCAAGGCAACAAAAGAGCAACTGAAAGCTGTTATGGATGATTTCGCAGCTTTTGTAGAGAAGTGCTGCAAGGCTGACGATAAGGAGACCTGCTTTGCCGAGGAGGGTAAAAAACTTGTTGCTGCAAGTCAAGCTGCCTTAGGCTTATAACATCACATTTAAAAGCATCTCAGCCTACCATGAGAATAAGAGAAAGAAAATGAAGATCAAAAGCTTATTCATCTGTTTTTCTTTTTCGTTGGTGTAAAGCCAACACCCTGTCTAAAAAACATAAATTTCTTTAATCATTTTGCCTCTTTTCTCTGTGCTTCAATTAATAAAAAA start: 234 end: 1242
Align Title:gi|6650825|gb|AF118090.1|AF118090 Homo sapiens PRO2044 mRNA, complete cds
Align Len: 1759
Score: 1983.0 Bits: 1789.33 E-val: 0.0
N.aligns:None Ident:996 Pos.:996 Gaps:0 Align len:999
Strand: (None, None) Frame: (1, 1)
Query: CAAAAACTATGCTGAGGCAAAGGATGTCTTCCTGGGCATGTTTTTGTATGAATATGCAAGAAGGCATCCTGATTACTCTGTCGTGCTGCTGCTGAGACTTGCCAAGACATATAAAACCACTCTAGAGAAGTGCTGTGCCGCTGCAGATCCTCATGAATGCTATGCCAAAGTGTTCGATGAATTTAAACCTCTTGTGGAAGAGCCTCAGAATTTAATCAAACAAAATTGTGAGCTTTTTGAGCAGCTTGGAGAGTACAAATTCCAGAATGCGCTATTAGTTCGTTACACCAAGAAAGTACCCCAAGTGTCAACTCCAACTCTTGTAGAGGTCTCAAGAAACCTAGGAAAAGTGGGCAGCAAATGTTGTAAACATCCTGAAGCAAAAAGAATGCCCTGTGCAGAAGACTATCTATCCGTGGTCCTGAACCAGTTATGTGTGTTGCATGAGAAAACGCCAGTAAGTGACAGAGTCACCAAATGCTGCACAGAATCCTTGGTGAACAGGCGACCATGCTTTTCAGCTCTGGAAGTCGATGAAACATACGTTCCCAAAGAGTTTAATGCTGAAACATTCACCTTCCATGCAGATATATGCACACTTTCTGAGAAGGAGAGACAAATCAAGAAACAAACTGCACTTGTTGAGCTTGTGAAACACAAGCCCAAGGCAACAAAAGAGCAACTGAAAGCTGTTATGGATGATTTCGCAGCTTTTGTAGAGAAGTGCTGCAAGGCTGACGATAAGGAGACCTGCTTTGCCGAGGAGGGTAAAAAACTTGTTGCTGCAAGTCGAGCTGCCTTAGGCTTATAACATCACATTTAAAAGCATCTCAGCCTACCATGAGAATAAGAGAAAGAAAATGAAGATCAAAAGCTTATTCATCTGTTTTTCTTTTTCGTTGGTGTAAAGCCAACACCCTGTCTAAAAAACATAAATTTCTTTAATCATTTTGCCTCTTTTCTCTGTGCTTCAATTAATAAAAAATGGAAAGAATCTAA start: 1053 end: 2051
Match: |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Subjc: CAAAAACTATGCTGAGGCAAAGGATGTCTTCCTGGGCATGTTTTTGTATGAATATGCAAGAAGGCATCCTGATTACTCTGTCGTGCTGCTGCTGAGACTTGCCAAGACATATGAAACCACTCTAGAGAAGTGCTGTGCCGCTGCAGATCCTCATGAATGCTATGCCAAAGTGTTCGATGAATTTAAACCTCTTGTGGAAGAGCCTCAGAATTTAATCAAACAAAATTGTGAGCTTTTTGAGCAGCTTGGAGAGTACAAATTCCAGAATGCGCTATTAGTTCGTTACACCAAGAAAGTACCCCAAGTGTCAACTCCAACTCTTGTAGAGGTCTCAAGAAACCTAGGAAAAGTGGGCAGCAAATGTTGTAAACATCCTGAAGCAAAAAGAATGCCCTGTGCAGAAGACTATCTATCCGTGGTCCTGAACCAGTTATGTGTGTTGCATGAGAAAACGCCAGTAAGTGACAGAGTCACCAAATGCTGCACAGAATCCTTGGTGAACAGGCGACCATGCTTTTCAGCTCTGGAAGTCGATGAAACATACGTTCCCAAAGAGTTTAATGCTGAAACATTCACCTTCCATGCAGATATATGCACACTTTCTGAGAAGGAGAGACAAATCAAGAAACAAACTGCACTTGTTGAGCTCGTGAAACACAAGCCCAAGGCAACAAAAGAGCAACTGAAAGCTGTTATGGATGATTTCGCAGCTTTTGTAGAGAAGTGCTGCAAGGCTGACGATAAGGAGACCTGCTTTGCCGAGGAGGGTAAAAAACTTGTTGCTGCAAGTCAAGCTGCCTTAGGCTTATAACATCACATTTAAAAGCATCTCAGCCTACCATGAGAATAAGAGAAAGAAAATGAAGATCAAAAGCTTATTCATCTGTTTTTCTTTTTCGTTGGTGTAAAGCCAACACCCTGTCTAAAAAACATAAATTTCTTTAATCATTTTGCCTCTTTTCTCTGTGCTTCAATTAATAAAAAATGGAAAGAATCTAA start: 692 end: 1690
Align Title:gi|168745965|emb|CU691683.1| Synthetic construct Homo sapiens gateway clone IMAGE:100022016 3' read mRNA
Align Len: 1128
Score: 1951.0 Bits: 1760.47 E-val: 0.0
N.aligns:None Ident:1034 Pos.:1034 Gaps:9 Align len:1061
Strand: (None, None) Frame: (1, -1)
Query: TGTGCTGATGACAGGGCGGACCTTGCCAAGTATAT-CTGTGAAAATCAAGATT-CGATCTCCAGTAAACT-GAAGGAATGCTGTGAAAAACCTCTGTTGGAAAAAT-CCCACTGCATTGCCGAAGTGGAAAATGATGAGATGCCTGCTGACTTGCCTTCATTAGCTGCTGATTTTGTTGAAAGTAA-GGATGTTTGCAAAAA-CTATGCTGAGGCAAAGGATGTCTTCCTGGGCATGTTTTTGTATGAATATGCAAGAAGGCATCCTGATTACTCTGTCGTGCTGCTGCTGAGACTTGCCAAGACATATAAAACCACTCTAGAGAAGTGCTGTGCCGCTGCAGATCCTCATGAATGCTATGCCAAAGTGTTCGATGAATTTAAACCTCTTGTGGAAGAGCCTCAGAATTTAATCAAACAAAATTGTGAGCTTTTTGAGCAGCTTGGAGAGTACAAATTCCAGAATGCGCTATTAGTTCGTTACACCAAGAAAGTACCCCAAGTGTCAACTCCAACTCTTGTAGAGGTCTCAAGAAACCTAGGAAAAGTGGGCAGCAAATGTTGTAAACATCCTGAAGCAAAAAGAATGCCCTGTGCAGAAGACTATCTATCCGTGGTCCTGAACCAGTTATGTGTGTTGCATGAGAAAACGCCAGTAAGTGACAGAGTCACCAAATGCTGCACAGAATCCTTGGTGAACAGGCGACCATGCTTTTCAGCTCTGGAAGTCGATGAAACATACGTTCCCAAAGAGTTTAATGCTGAAACATTCACCTTCCATGCAGATATATGCACACTTTCTGAGAAGGAGAGACAAATCAAGAAACAAACTGCACTTGTTGAGCTTGTGAAACACAAGCCCAAGGCAACAAAAGAGCAACTGAAAGCTGTTATGGATGATTTCGCAGCTTTTGTAGAGAAGTGCTGCAAGGCTGACGATAAGGAGACCTGCTTTGCCGAGGAGGGTAA-AAAACTTGTTGCTGCAAGTCGAGCTGCCTTAGGCTTATAACATCACATTTAAAAGCATCTCAGCCTACCATGAGAATAAGAGAAAGAAAATG start: 862 end: 1915
Match: || ||||| ||||||| ||||||||||||| | | ||| |||||||||||| |||| ||||||||||| |||||||||| ||||||||||| ||||||||||| ||||||||||||| |||||||||||||||||||||| |||||||| | |||||||||||||||||||||||||||||| ||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||| ||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Subjc: TGGGCTGAGAACAGGGCCGACCTTGCCAAGTTTTTTCTGGAAAAATCAAGATTTCGATTTCCAGTAAACTAGAAGGAATGC-GTGAAAAACCTTCGTTGGAAAAATTCCCACTGCATTGC-GAAGTGGAAAATGATGAGATGCTTGCTGACTGCCTTTCATTAGCTGCTGATTTTGTTGAAAGTAAAGGATGTTTGCAAAAAACTATGCTGAGGCAAAGGATGTCTTCCTGGGCATGTTTTTGTATGAATATGCAAGAAAGCATCCTGATTACTCTGTCGTGCTGCTGCTGAGACTTGCCAAGACATATGAAACCACTCTAGAGAAGTGCTGTGCCGCTGCAGATCCTCATGAATGCTATGCCAAAGTGTTCGATGAATTTAAACCTCTTGTGGAAGAGCCTCAGAATTTAATCAAACAAAATTGTGAGCTTTTTGAGCAGCTTGGAGAGTACAAATTCCAGAATGCGCTATTAGTTCGTTACACCAAGAAAGTACCCCAAGTGTCAACTCCAACTCTTGTAGAGGTCTCAAGAAACCTAGGAAAAGTGGGCAGCAAATGTTGTAAACATCCTGAAGCAAAAAGAATGCCCTGTGCAGAAGACTATCTATCCGTGGTCCTGAACCAGTTATGTGTGTTGCATGAGAAAACGCCAGTAAGTGACAGAGTCACCAAATGCTGCACAGAATCCTTGGTGAACAGGCGACCATGCTTTTCAGCTCTGGAAGTCGATGAAACATACGTTCCCAAAGAGTTTAATGCTGAAACATTCACCTTCCATGCAGATATATGCACACTTTCTGAGAAGGAGAGACAAATCAAGAAACAAACTGCACTTGTTGAGCTTGTGAAACACAAGCCCAAGGCAACAAAAGAGCAACTGAAAGCTGTTATGGATGATTTCGCAGCTTTTGTAGAGAAGTGCTGCAAGGCTGACGATAAGGAGACCTGCTTTGCCGAGGAGGGTAAAAAAACTTGTTGCTGCAAGTCAAGCTGCCTTAGGCTTATAACATCACATTTAAAAGCATCTCAGCCTACCATGAGAATAAGAGAAAGAAAATG start: 1075 end: 17
Align Title:gi|168745964|emb|CU691682.1| Synthetic construct Homo sapiens gateway clone IMAGE:100022016 5' read mRNA
Align Len: 1251
Score: 1896.0 Bits: 1710.88 E-val: 0.0
N.aligns:None Ident:1082 Pos.:1082 Gaps:15 Align len:1153
Strand: (None, None) Frame: (1, 1)
Query: ATGAAGTGGGTAACCTTTATTTCCCTTCTTTTTCTCTTTAGCTCGGCTTATTCCAGGGGTGTGTTTCGTCGAGATGCACACAAGAGTGAGGTTGCTCATCGGTTTAAAGATTTGGGAGAAGAAAATTTCAAAGCCTTAGTGTTGATTGCCTTTGCTCAGTATCTTCAGCAGTGTCCATTTGAAGATCATGTAAAATTAGTGAATGAAGTAACTGAATTTGCAAAAACATGTGTTGCTGATGAGTCAGCTGAAAATTGTGACAAATCACTTCATACCCTTTTTGGAGACAAATTATGCACAGTTGCAACTCTTCGTGAAACCTATGGTGAAATGGCTGACTGCTGTGCAAAACAAGAACCTGAGAGAAATGAATGCTTCTTGCAACACAAAGATGACAACCCAAACCTCCCCCGATTGGTGAGACCAGAGGTTGATGTGATGTGCACTGCTTTTCATGACAATGAAGAGACATTTTTGAAAAAATACTTATATGAAATTGCCAGAAGACATCCTTACTTCTATGCCCCGGAACTCCTTTTCTTTGCTAAAAGGTATAAAGCTGCTTTTACAGAATGTTGCCAAGCTGCTGATAAGGCTGCCTGCCTGTTGCCAAAGCTCGATGAACTTCGGGATGAAGGGAAGGCTTCGTCTGCCAAACAGAGGCTCAAGTGTGCCAGTCTCCAAAAA-TTTGGAGAAAGAGCTTTCAAAGCATGGGCAGTAGCTCGCCTGAGCCAGAGATTTCCCAAAGCTGAGTTTGCAGAAGTTTCCAAGTTAGTGACAGATCTTACCAAAGTCCACACGGAATGCTGCCATGGAGATCTGCTTGAATGTGCTGATGACAGGGCGGACCTTGCCAAGTATATCTGTGAAAATCAAGATTCGATCTCCAGTAAACTGAAGGAATGCTGTGAAAAACCTCTGTTGGAAAAATCCCACTGCATTGCCGAAGTGG-AAAATGATGAGATGCCTGCTGACTTGCCTTCATTAGCTGCTGATTTTGTTGAAAGTAAGGATGTTTGCAAAAACTATGCTGAGGCAAAGGATGTCTTCCTGGGCATGTTTTTGTATGAATATGCAAGAAGGCATCCTGATTACTCTGTCGTGCTGCTGCTGAGACTTGCCAAGACATATAAAACCACTCTAGAGAAGTG start: 34 end: 1184
Match: ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||| |||||||||||||||||| |||||||||||||||||||||||||||||||| ||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||| ||||||||||||||||||||||||||||| |||||||||| ||||||||||||||||||||||||||| |||||||||||| || |||||||||||||||||||||||||||||||||||| || | ||||| |||||| |||||||||| ||| ||| ||| || | || ||| ||||||| |||||| ||| |||||||||| ||||||| | | ||||||| || |||| |||||| ||||||| ||||| | ||| |||| |||| |||| | |||| | |||| | || | |||| | |||| | ||||| ||| | ||| |||
Subjc: ATGAAGTGGGTAACCTTTATTTCCCTTCTTTTTCTCTTTAGCTCGGCTTATTCCAGGGGTGTGTTTCGTCGAGATGCACACAAGAGTGAGGTTGCTCATCGGTTTAAAGATTTGGGAGAAGAAAATTTCAAAGCCTTGGTGTTGATTGCCTTTGCTCAGTATCTTCAGCAGTGTCCATTTGAAGATCATGTAAAATTAGTGAATGAAGTAACTGAATTTGCAAAAACATGTGTTGCTGATGAGTCAGCTGAAAATTGTGACAAATCACTTCATACCCTTTTTGGAGACAAATTATGCACAGTTGCAACTCTTCGTGAAACCTATGGTGAAATGGCTGACTGCTGTGCAAAACAAGAACCTGAGAGAAATGAATGCTTCTTGCAACACAAAGATGACAACCCAAACCTCCCCCGATTGGTGAGACCAGAGGTTGATGTGATGTGCACTGCTTTTCATGACAATGAAGAGACATTTTTGAAAAAATACTTATATGAAATTGCCAGAAGACATCCTTACTTTTATGCCCCGGAACTCCTTTTCTTTGCTAAAAGGTATAAAGCTGCTTTTACAGAATGTTGCCAAGCTGCTGATAAAGCTGCCTGCCTGTTGCCAAAGCTCGATGAACTTCGGGATGAAGGGAAGGCTTCGTCTGCCAAACAGAGACTCAAGTGTGCCAGTCTCCAAAAATTTTGGAGAAAGAGCTTTC-AAGCATGGGCAGTAGCTCGCCTGAGCCAGAGA-TTCCCAAAGCTGAGTTTGCAGAAGTTTCC-AGTTAGTGACAGATCTTACCAAAGTCCACACGGAATGCTGCCCTGGAGATCTGCTTGAATGTGCTGATGACAAGGCGGACCTTTGCAAGTATATCTGTGAAAATCAAGATTCAATCTCCAGTAAA-TGGAGGAATGCTGTGAAAAACCTCTGTTGGAAAAATCCCCCTTCCTTGCCCAAGTGGAAAAATGATGAAATG-CTGGTGAATTTGCCTCCTTAACTGCTGAATTTGTTTGAAGATAGGATGTTTG-AAAAACTTAGTTCAGGCAAAAGAGTTCTT-CTGGGCGATTTTTTGT-TGAATTTCCAA-AAGGGATCCGGATT-CCCTGTGGGGCTGGGGGGGA-ATTTGC--AAACATTTTAAACCTCTCCAAAGAGGTG start: 17 end: 1156
Align Title:gi|23241674|gb|BC035969.1| Homo sapiens albumin, mRNA (cDNA clone MGC:32581 IMAGE:4714468), complete cds
Align Len: 1449
Score: 1767.0 Bits: 1594.56 E-val: 0.0
N.aligns:None Ident:888 Pos.:888 Gaps:0 Align len:891
Strand: (None, None) Frame: (1, 1)
Query: ATATAAAACCACTCTAGAGAAGTGCTGTGCCGCTGCAGATCCTCATGAATGCTATGCCAAAGTGTTCGATGAATTTAAACCTCTTGTGGAAGAGCCTCAGAATTTAATCAAACAAAATTGTGAGCTTTTTGAGCAGCTTGGAGAGTACAAATTCCAGAATGCGCTATTAGTTCGTTACACCAAGAAAGTACCCCAAGTGTCAACTCCAACTCTTGTAGAGGTCTCAAGAAACCTAGGAAAAGTGGGCAGCAAATGTTGTAAACATCCTGAAGCAAAAAGAATGCCCTGTGCAGAAGACTATCTATCCGTGGTCCTGAACCAGTTATGTGTGTTGCATGAGAAAACGCCAGTAAGTGACAGAGTCACCAAATGCTGCACAGAATCCTTGGTGAACAGGCGACCATGCTTTTCAGCTCTGGAAGTCGATGAAACATACGTTCCCAAAGAGTTTAATGCTGAAACATTCACCTTCCATGCAGATATATGCACACTTTCTGAGAAGGAGAGACAAATCAAGAAACAAACTGCACTTGTTGAGCTTGTGAAACACAAGCCCAAGGCAACAAAAGAGCAACTGAAAGCTGTTATGGATGATTTCGCAGCTTTTGTAGAGAAGTGCTGCAAGGCTGACGATAAGGAGACCTGCTTTGCCGAGGAGGGTAAAAAACTTGTTGCTGCAAGTCGAGCTGCCTTAGGCTTATAACATCACATTTAAAAGCATCTCAGCCTACCATGAGAATAAGAGAAAGAAAATGAAGATCAAAAGCTTATTCATCTGTTTTTCTTTTTCGTTGGTGTAAAGCCAACACCCTGTCTAAAAAACATAAATTTCTTTAATCATTTTGCCTCTTTTCTCTGTGCTTCAATTAATAAAAAATGGAAAGAATCTAA start: 1161 end: 2051
Match: |||| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Subjc: ATATGAAACCACTCTAGAGAAGTGCTGTGCCGCTGCAGATCCTCATGAATGCTATGCCAAAGTGTTCGATGAATTTAAACCTCTTGTGGAAGAGCCTCAGAATTTAATCAAACAAAATTGTGAGCTTTTTGAGCAGCTTGGAGAGTACAAATTCCAGAATGCGCTATTAGTTCGTTACACCAAGAAAGTACCCCAAGTGTCAACTCCAACTCTTGTAGAGGTCTCAAGAAACCTAGGAAAAGTGGGCAGCAAATGTTGTAAACATCCTGAAGCAAAAAGAATGCCCTGTGCAGAAGACTATCTATCCGTGGTCCTGAACCAGTTATGTGTGTTGCATGAGAAAACGCCAGTAAGTGACAGAGTCACCAAATGCTGCACAGAATCCTTGGTGAACAGGCGACCATGCTTTTCAGCTCTGGAAGTCGATGAAACATACGTTCCCAAAGAGTTTAATGCTGAAACATTCACCTTCCATGCAGATATATGCACACTTTCTGAGAAGGAGAGACAAATCAAGAAACAAACTGCACTTGTTGAGCTCGTGAAACACAAGCCCAAGGCAACAAAAGAGCAACTGAAAGCTGTTATGGATGATTTCGCAGCTTTTGTAGAGAAGTGCTGCAAGGCTGACGATAAGGAGACCTGCTTTGCCGAGGAGGGTAAAAAACTTGTTGCTGCAAGTCAAGCTGCCTTAGGCTTATAACATCACATTTAAAAGCATCTCAGCCTACCATGAGAATAAGAGAAAGAAAATGAAGATCAAAAGCTTATTCATCTGTTTTTCTTTTTCGTTGGTGTAAAGCCAACACCCTGTCTAAAAAACATAAATTTCTTTAATCATTTTGCCTCTTTTCTCTGTGCTTCAATTAATAAAAAATGGAAAGAATCTAA start: 530 end: 1420
Score: 1048.0 Bits: 946.252 E-val: 0.0
N.aligns:None Ident:527 Pos.:527 Gaps:0 Align len:529
Strand: (None, None) Frame: (1, 1)
Query: TCTCTTCTGTCAACCCCACGCGCCTTTGGCACAATGAAGTGGGTAACCTTTATTTCCCTTCTTTTTCTCTTTAGCTCGGCTTATTCCAGGGGTGTGTTTCGTCGAGATGCACACAAGAGTGAGGTTGCTCATCGGTTTAAAGATTTGGGAGAAGAAAATTTCAAAGCCTTAGTGTTGATTGCCTTTGCTCAGTATCTTCAGCAGTGTCCATTTGAAGATCATGTAAAATTAGTGAATGAAGTAACTGAATTTGCAAAAACATGTGTTGCTGATGAGTCAGCTGAAAATTGTGACAAATCACTTCATACCCTTTTTGGAGACAAATTATGCACAGTTGCAACTCTTCGTGAAACCTATGGTGAAATGGCTGACTGCTGTGCAAAACAAGAACCTGAGAGAAATGAATGCTTCTTGCAACACAAAGATGACAACCCAAACCTCCCCCGATTGGTGAGACCAGAGGTTGATGTGATGTGCACTGCTTTTCATGACAATGAAGAGACATTTTTGAAAAAATACTTATATGAAA start: 1 end: 529
Match: ||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Subjc: TCTCTTCTGTCAACCCCACACGCCTTTGGCACAATGAAGTGGGTAACCTTTATTTCCCTTCTTTTTCTCTTTAGCTCGGCTTATTCCAGGGGTGTGTTTCGTCGAGATGCACACAAGAGTGAGGTTGCTCATCGGTTTAAAGATTTGGGAGAAGAAAATTTCAAAGCCTTGGTGTTGATTGCCTTTGCTCAGTATCTTCAGCAGTGTCCATTTGAAGATCATGTAAAATTAGTGAATGAAGTAACTGAATTTGCAAAAACATGTGTTGCTGATGAGTCAGCTGAAAATTGTGACAAATCACTTCATACCCTTTTTGGAGACAAATTATGCACAGTTGCAACTCTTCGTGAAACCTATGGTGAAATGGCTGACTGCTGTGCAAAACAAGAACCTGAGAGAAATGAATGCTTCTTGCAACACAAAGATGACAACCCAAACCTCCCCCGATTGGTGAGACCAGAGGTTGATGTGATGTGCACTGCTTTTCATGACAATGAAGAGACATTTTTGAAAAAATACTTATATGAAA start: 9 end: 537
Align Title:gi|15679995|gb|BC014308.1|BC014308 Homo sapiens, clone IMAGE:3934797, mRNA, partial cds
Align Len: 823
Score: 1571.0 Bits: 1417.83 E-val: 0.0
N.aligns:None Ident:790 Pos.:790 Gaps:0 Align len:793
Strand: (None, None) Frame: (1, 1)
Query: AGAATTTAATCAAACAAAATTGTGAGCTTTTTGAGCAGCTTGGAGAGTACAAATTCCAGAATGCGCTATTAGTTCGTTACACCAAGAAAGTACCCCAAGTGTCAACTCCAACTCTTGTAGAGGTCTCAAGAAACCTAGGAAAAGTGGGCAGCAAATGTTGTAAACATCCTGAAGCAAAAAGAATGCCCTGTGCAGAAGACTATCTATCCGTGGTCCTGAACCAGTTATGTGTGTTGCATGAGAAAACGCCAGTAAGTGACAGAGTCACCAAATGCTGCACAGAATCCTTGGTGAACAGGCGACCATGCTTTTCAGCTCTGGAAGTCGATGAAACATACGTTCCCAAAGAGTTTAATGCTGAAACATTCACCTTCCATGCAGATATATGCACACTTTCTGAGAAGGAGAGACAAATCAAGAAACAAACTGCACTTGTTGAGCTTGTGAAACACAAGCCCAAGGCAACAAAAGAGCAACTGAAAGCTGTTATGGATGATTTCGCAGCTTTTGTAGAGAAGTGCTGCAAGGCTGACGATAAGGAGACCTGCTTTGCCGAGGAGGGTAAAAAACTTGTTGCTGCAAGTCGAGCTGCCTTAGGCTTATAACATCACATTTAAAAGCATCTCAGCCTACCATGAGAATAAGAGAAAGAAAATGAAGATCAAAAGCTTATTCATCTGTTTTTCTTTTTCGTTGGTGTAAAGCCAACACCCTGTCTAAAAAACATAAATTTCTTTAATCATTTTGCCTCTTTTCTCTGTGCTTCAATTAATAAAAAATGGAAAGAATCTAA start: 1259 end: 2051
Match: |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| ||
Subjc: AGAATTTAATCAAACAAAATTGTGAGCTTTTTGAGCAGCTTGGAGAGTACAAATTCCAGAATGCGCTATTAGTTCGTTACACCAAGAAAGTACCCCAAGTGTCAACTCCAACTCTTGTAGAGGTCTCAAGAAACCTAGGAAAAGTGGGCAGCAAATGTTGTAAACATCCTGAAGCAAAAAGAATGCCCTGTGCAGAAGACTATCTATCCGTGGTCCTGAACCAGTTATGTGTGTTGCATGAGAAAACGCCAGTAAGTGACAGAGTCACCAAATGCTGCACAGAATCCTTGGTGAACAGGCGACCATGCTTTTCAGCTCTGGAAGTCGATGAAACATACGTTCCCAAAGAGTTTAATGCTGAAACATTCACCTTCCATGCAGATATATGCACACTTTCTGAGAAGGAGAGACAAATCAAGAAACAAACTGCACTTGTTGAGCTCGTGAAACACAAGCCCAAGGCAACAAAAGAGCAACTGAAAGCTGTTATGGATGATTTCGCAGCTTTTGTAGAGAAGTGCTGCAAGGCTGACGATAAGGAGACCTGCTTTGCCGAGGAGGGTAAAAAACTTGTTGCTGCAAGTCAAGCTGCCTTAGGCTTATAACATCACATTTAAAAGCATCTCAGCCTACCATGAGAATAAGAGAAAGAAAATGAAGATCAAAAGCTTATTCATCTGTTTTTCTTTTTCGTTGGTGTAAAGCCAACACCCTGTCTAAAAAACATAAATTTCTTTAATCATTTTGCCTCTTTTCTCTGTGCTTCAATTAATAAAAAATGGAAAGAATCCAA start: 4 end: 796
Align Title:gi|649152310|gb|KJ905683.1| Synthetic construct Homo sapiens clone ccsbBroadEn_15353 ALB gene, encodes complete protein
Align Len: 1320
Score: 1387.0 Bits: 1251.92 E-val: 0.0
N.aligns:None Ident:698 Pos.:698 Gaps:0 Align len:701
Strand: (None, None) Frame: (1, 1)
Query: ATATAAAACCACTCTAGAGAAGTGCTGTGCCGCTGCAGATCCTCATGAATGCTATGCCAAAGTGTTCGATGAATTTAAACCTCTTGTGGAAGAGCCTCAGAATTTAATCAAACAAAATTGTGAGCTTTTTGAGCAGCTTGGAGAGTACAAATTCCAGAATGCGCTATTAGTTCGTTACACCAAGAAAGTACCCCAAGTGTCAACTCCAACTCTTGTAGAGGTCTCAAGAAACCTAGGAAAAGTGGGCAGCAAATGTTGTAAACATCCTGAAGCAAAAAGAATGCCCTGTGCAGAAGACTATCTATCCGTGGTCCTGAACCAGTTATGTGTGTTGCATGAGAAAACGCCAGTAAGTGACAGAGTCACCAAATGCTGCACAGAATCCTTGGTGAACAGGCGACCATGCTTTTCAGCTCTGGAAGTCGATGAAACATACGTTCCCAAAGAGTTTAATGCTGAAACATTCACCTTCCATGCAGATATATGCACACTTTCTGAGAAGGAGAGACAAATCAAGAAACAAACTGCACTTGTTGAGCTTGTGAAACACAAGCCCAAGGCAACAAAAGAGCAACTGAAAGCTGTTATGGATGATTTCGCAGCTTTTGTAGAGAAGTGCTGCAAGGCTGACGATAAGGAGACCTGCTTTGCCGAGGAGGGTAAAAAACTTGTTGCTGCAAGTCGAGCTGCCTTAGGCTTAT start: 1161 end: 1861
Match: |||| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||
Subjc: ATATGAAACCACTCTAGAGAAGTGCTGTGCCGCTGCAGATCCTCATGAATGCTATGCCAAAGTGTTCGATGAATTTAAACCTCTTGTGGAAGAGCCTCAGAATTTAATCAAACAAAATTGTGAGCTTTTTGAGCAGCTTGGAGAGTACAAATTCCAGAATGCGCTATTAGTTCGTTACACCAAGAAAGTACCCCAAGTGTCAACTCCAACTCTTGTAGAGGTCTCAAGAAACCTAGGAAAAGTGGGCAGCAAATGTTGTAAACATCCTGAAGCAAAAAGAATGCCCTGTGCAGAAGACTATCTATCCGTGGTCCTGAACCAGTTATGTGTGTTGCATGAGAAAACGCCAGTAAGTGACAGAGTCACCAAATGCTGCACAGAATCCTTGGTGAACAGGCGACCATGCTTTTCAGCTCTGGAAGTCGATGAAACATACGTTCCCAAAGAGTTTAATGCTGAAACATTCACCTTCCATGCAGATATATGCACACTTTCTGAGAAGGAGAGACAAATCAAGAAACAAACTGCACTTGTTGAGCTCGTGAAACACAAGCCCAAGGCAACAAAAGAGCAACTGAAAGCTGTTATGGATGATTTCGCAGCTTTTGTAGAGAAGTGCTGCAAGGCTGACGATAAGGAGACCTGCTTTGCCGAGGAGGGTAAAAAACTTGTTGCTGCAAGTCAAGCTGCCTTAGGCTTAT start: 554 end: 1254
Score: 987.0 Bits: 891.249 E-val: 0.0
N.aligns:None Ident:495 Pos.:495 Gaps:0 Align len:496
Strand: (None, None) Frame: (1, 1)
Query: ATGAAGTGGGTAACCTTTATTTCCCTTCTTTTTCTCTTTAGCTCGGCTTATTCCAGGGGTGTGTTTCGTCGAGATGCACACAAGAGTGAGGTTGCTCATCGGTTTAAAGATTTGGGAGAAGAAAATTTCAAAGCCTTAGTGTTGATTGCCTTTGCTCAGTATCTTCAGCAGTGTCCATTTGAAGATCATGTAAAATTAGTGAATGAAGTAACTGAATTTGCAAAAACATGTGTTGCTGATGAGTCAGCTGAAAATTGTGACAAATCACTTCATACCCTTTTTGGAGACAAATTATGCACAGTTGCAACTCTTCGTGAAACCTATGGTGAAATGGCTGACTGCTGTGCAAAACAAGAACCTGAGAGAAATGAATGCTTCTTGCAACACAAAGATGACAACCCAAACCTCCCCCGATTGGTGAGACCAGAGGTTGATGTGATGTGCACTGCTTTTCATGACAATGAAGAGACATTTTTGAAAAAATACTTATATGAAA start: 34 end: 529
Match: ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Subjc: ATGAAGTGGGTAACCTTTATTTCCCTTCTTTTTCTCTTTAGCTCGGCTTATTCCAGGGGTGTGTTTCGTCGAGATGCACACAAGAGTGAGGTTGCTCATCGGTTTAAAGATTTGGGAGAAGAAAATTTCAAAGCCTTGGTGTTGATTGCCTTTGCTCAGTATCTTCAGCAGTGTCCATTTGAAGATCATGTAAAATTAGTGAATGAAGTAACTGAATTTGCAAAAACATGTGTTGCTGATGAGTCAGCTGAAAATTGTGACAAATCACTTCATACCCTTTTTGGAGACAAATTATGCACAGTTGCAACTCTTCGTGAAACCTATGGTGAAATGGCTGACTGCTGTGCAAAACAAGAACCTGAGAGAAATGAATGCTTCTTGCAACACAAAGATGACAACCCAAACCTCCCCCGATTGGTGAGACCAGAGGTTGATGTGATGTGCACTGCTTTTCATGACAATGAAGAGACATTTTTGAAAAAATACTTATATGAAA start: 66 end: 561
Align Title:gi|168740012|emb|CU690239.1| Synthetic construct Homo sapiens gateway clone IMAGE:100022252 3' read ALB mRNA
Align Len: 1356
Score: 1182.0 Bits: 1067.08 E-val: 0.0
N.aligns:None Ident:647 Pos.:647 Gaps:12 Align len:670
Strand: (None, None) Frame: (1, -1)
Query: CATGAATG-CTAT--GCCAAA-GTGTT-CGATG-AATTTAAA-CCTCTTGTGG-AAGAGCCTCAG-AATTTAAT-CAAACAAAATT-GTGAGCTTTTTGAGCAGCTTGGAGAGTACAAATTCCAGAATGCGCTATTAGTTCGTTACACCAAGAAAGTACCCCAAGTGTCAACTCCAACTCTTGTAGAGGTCTCAAGAAACCTAGGAAAAGTGGGCAGCAAATGTTGTAAACATCCTGAAGCAAAAAGAATGCCCTGTGCAGAAGACTATCTATCCGTGGTCCTGAACCAGTTATGTGTGTTGCATGAGAAAACGCCAGTAAGTGACAGAGTCACCAAATGCTGCACAGAATCCTTGGTGAACAGGCGACCATGCTTTTCAGCTCTGGAAGTCGATGAAACATACGTTCCCAAAGAGTTTAATGCTGAAACATTCACCTTCCATGCAGATATATGCACACTTTCTGAGAAGGAGAGACAAATCAAGAAACAAACTGCACTTGTTGAGCTTGTGAAACACAAGCCCAAGGCAACAAAAGAGCAACTGAAAGCTGTTATGGATGATTTCGCAGCTTTTGTAGAGAAGTGCTGCAAGGCTGACGATAAGGAGACCTGCTTTGCCGAGGAGGGTAAAAAACTTGTTGCTGCAAGTCGAGCTGCCTTAGGCTTATA start: 1204 end: 1862
Match: |||||||| ||| |||||| ||||| ||||| |||||||| ||| |||||| ||||||||||| |||||||| ||||||||||| |||||||||||| ||||||||||||||||| | |||||||||||||| ||||||| ||||| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| ||||||||||||||||||
Subjc: CATGAATGACTAATGGCCAAAAGTGTTTCGATGGAATTTAAAACCTTTTGTGGTAAGAGCCTCAGGAATTTAATTCAAACAAAATTTGTGAGCTTTTTGGCCAGCTTGGAGAGTACAATTCCCAGAATGCGCTATAAGTTCGT-ACACCCTGAAAGTACCCCAAGTGTCAACTCCAACTCTTGTAGAGGTCTCAAGAAACCTAGGAAAAGTGGGCAGCAAATGTTGTAAACATCCTGAAGCAAAAAGAATGCCCTGTGCAGAAGACTATCTATCCGTGGTCCTGAACCAGTTATGTGTGTTGCATGAGAAAACGCCAGTAAGTGACAGAGTCACCAAATGCTGCACAGAATCCTTGGTGAACAGGCGACCATGCTTTTCAGCTCTGGAAGTCGATGAAACATACGTTCCCAAAGAGTTTAATGCTGAAACATTCACCTTCCATGCAGATATATGCACACTTTCTGAGAAGGAGAGACAAATCAAGAAACAAACTGCACTTGTTGAGCTCGTGAAACACAAGCCCAAGGCAACAAAAGAGCAACTGAAAGCTGTTATGGATGATTTCGCAGCTTTTGTAGAGAAGTGCTGCAAGGCTGACGATAAGGAGACCTGCTTTGCCGAGGAGGGTAAAAAACTTGTTGCTGCAAGTCAAGCTGCCTTAGGCTTATA start: 685 end: 17
Align Title:gi|37181745|gb|AY358313.1| Homo sapiens DNA66677 ALB (UNQ696) mRNA, partial cds
Align Len: 524
Score: 1018.0 Bits: 919.201 E-val: 0.0
N.aligns:None Ident:517 Pos.:517 Gaps:0 Align len:523
Strand: (None, None) Frame: (1, 1)
Query: AACCCCACGCGCCTTTGGCACAATGAAGTGGGTAACCTTTATTTCCCTTCTTTTTCTCTTTAGCTCGGCTTATTCCAGGGGTGTGTTTCGTCGAGATGCACACAAGAGTGAGGTTGCTCATCGGTTTAAAGATTTGGGAGAAGAAAATTTCAAAGCCTTAGTGTTGATTGCCTTTGCTCAGTATCTTCAGCAGTGTCCATTTGAAGATCATGTAAAATTAGTGAATGAAGTAACTGAATTTGCAAAAACATGTGTTGCTGATGAGTCAGCTGAAAATTGTGACAAATCACTTCATACCCTTTTTGGAGACAAATTATGCACAGTTGCAACTCTTCGTGAAACCTATGGTGAAATGGCTGACTGCTGTGCAAAACAAGAACCTGAGAGAAATGAATGCTTCTTGCAACACAAAGATGACAACCCAAACCTCCCCCGATTGGTGAGACCAGAGGTTGATGTGATGTGCACTGCTTTTCATGACAATGAAGAGACATTTTTGAAAAAATACTTATATGAAATTGCC start: 12 end: 534
Match: ||||||| || || ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Subjc: AACCCCANANGCTTTGGGCACAATGAAGTGGGTAACCTTTATTTCCCTTCTTTTTCTCTTTAGCTCGGCTTATTCCAGGGGTGTGTTTCGTCGAGATGCACACAAGAGTGAGGTTGCTCATCGGTTTAAAGATTTGGGAGAAGAAAATTTCAAAGCCTTGGTGTTGATTGCCTTTGCTCAGTATCTTCAGCAGTGTCCATTTGAAGATCATGTAAAATTAGTGAATGAAGTAACTGAATTTGCAAAAACATGTGTTGCTGATGAGTCAGCTGAAAATTGTGACAAATCACTTCATACCCTTTTTGGAGACAAATTATGCACAGTTGCAACTCTTCGTGAAACCTATGGTGAAATGGCTGACTGCTGTGCAAAACAAGAACCTGAGAGAAATGAATGCTTCTTGCAACACAAAGATGACAACCCAAACCTCCCCCGATTGGTGAGACCAGAGGTTGATGTGATGTGCACTGCTTTTCATGACAATGAAGAGACATTTTTGAAAAAATACTTATATGAAATTGCC start: 2 end: 524
Align Title:gi|168740011|emb|CU690238.1| Synthetic construct Homo sapiens gateway clone IMAGE:100022252 5' read ALB mRNA
Align Len: 1442
Score: 982.0 Bits: 886.74 E-val: 0.0
N.aligns:None Ident:494 Pos.:494 Gaps:0 Align len:496
Strand: (None, None) Frame: (1, 1)
Query: ATGAAGTGGGTAACCTTTATTTCCCTTCTTTTTCTCTTTAGCTCGGCTTATTCCAGGGGTGTGTTTCGTCGAGATGCACACAAGAGTGAGGTTGCTCATCGGTTTAAAGATTTGGGAGAAGAAAATTTCAAAGCCTTAGTGTTGATTGCCTTTGCTCAGTATCTTCAGCAGTGTCCATTTGAAGATCATGTAAAATTAGTGAATGAAGTAACTGAATTTGCAAAAACATGTGTTGCTGATGAGTCAGCTGAAAATTGTGACAAATCACTTCATACCCTTTTTGGAGACAAATTATGCACAGTTGCAACTCTTCGTGAAACCTATGGTGAAATGGCTGACTGCTGTGCAAAACAAGAACCTGAGAGAAATGAATGCTTCTTGCAACACAAAGATGACAACCCAAACCTCCCCCGATTGGTGAGACCAGAGGTTGATGTGATGTGCACTGCTTTTCATGACAATGAAGAGACATTTTTGAAAAAATACTTATATGAAA start: 34 end: 529
Match: ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||
Subjc: ATGAAGTGGGTAACCTTTATTTCCCTTCTTTTTCTCTTTAGCTCGGCTTATTCCAGGGGTGTGTTTCGTCGAGATGCACACAAGAGTGAGGTTGCTCATCGGTTTAAAGATTTGGGAGAAGAAAATTTCAAAGCCTTGGTGTTGATTGCCTTTGCTCAGTATCTTCAGCAGTGTCCATTTGAAGATCATGTAAAATTAGTGAATGAAGTAACTGAATTTGCAAAAACATGTGTTGCTGATGAGTCAGCTGAAAATTGTGACAAATCACTTCATACCCTTTTTGGAGACAAATTATGCACAGTTGCAACTCTTCGTGAAACCTATGGTGAAATGGCTGACTGCTGTGCAAAACAAGAACCTGAGAGAAATGAATGCTTCTTGCAACACAAAGATGACAACCCAAACCTCCCCCGATTGGTGAGACCAGAGGTTGATGTGATGTGCACTGCTTTTCATGACAATGAAGAGACCTTTTTGAAAAAATACTTATATGAAA start: 17 end: 512
Score: 143.0 Bits: 130.227 E-val: 1.83659e-26
N.aligns:None Ident:122 Pos.:122 Gaps:13 Align len:145
Strand: (None, None) Frame: (1, 1)
Query: ATATAAAACCACTCTAGAGAA-GTGCTGTGCC-GCTGCAGATCCTCATGAATGCTA-TGCCAAAGTGTTCGATGAATTTAAACC--TCTTGTGGAA-GAGCCTCAGAA--TTTAATCAAAC---AAAATTG-TGAG-CTTTTTGA start: 1161 end: 1292
Match: |||| |||||||||||||||| |||||||||| ||||||||||||||||||||||| || || ||||| |||||||| |||||| |||| |||| ||||||||| | ||||||||||| | | ||| |||| ||||||||
Subjc: ATATGAAACCACTCTAGAGAAAGTGCTGTGCCCGCTGCAGATCCTCATGAATGCTATTGACATAGTGTACGATGAATCTAAACCCTCCTTGGGGAATGAGCCTCAGTACTTTTAATCAAACTCAACACTTGTTGAGCCTTTTTGA start: 505 end: 649
Align Title:gi|6642751|gb|AF113014.1|AF113014 Homo sapiens clone FLB3512 mRNA sequence
Align Len: 1476
Score: 618.0 Bits: 558.527 E-val: 1.15187e-155
N.aligns:None Ident:312 Pos.:312 Gaps:0 Align len:314
Strand: (None, None) Frame: (1, 1)
Query: AAAGCTGTTATGGATGATTTCGCAGCTTTTGTAGAGAAGTGCTGCAAGGCTGACGATAAGGAGACCTGCTTTGCCGAGGAGGGTAAAAAACTTGTTGCTGCAAGTCGAGCTGCCTTAGGCTTATAACATCACATTTAAAAGCATCTCAGCCTACCATGAGAATAAGAGAAAGAAAATGAAGATCAAAAGCTTATTCATCTGTTTTTCTTTTTCGTTGGTGTAAAGCCAACACCCTGTCTAAAAAACATAAATTTCTTTAATCATTTTGCCTCTTTTCTCTGTGCTTCAATTAATAAAAAATGGAAAGAATCTAA start: 1738 end: 2051
Match: ||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Subjc: AAAGCTGCTATGGATGATTTCGCAGCTTTTGTAGAGAAGTGCTGCAAGGCTGACGATAAGGAGACCTGCTTTGCCGAGGAGGGTAAAAAACTTGTTGCTGCAAGTCAAGCTGCCTTAGGCTTATAACATCACATTTAAAAGCATCTCAGCCTACCATGAGAATAAGAGAAAGAAAATGAAGATCAAAAGCTTATTCATCTGTTTTTCTTTTTCGTTGGTGTAAAGCCAACACCCTGTCTAAAAAACATAAATTTCTTTAATCATTTTGCCTCTTTTCTCTGTGCTTCAATTAATAAAAAATGGAAAGAATCTAA start: 1153 end: 1466
Align Title:gi|219842221|ref|NG_009291.1| Homo sapiens albumin (ALB), RefSeqGene on chromosome 4
Align Len: 24158
Score: 581.0 Bits: 525.165 E-val: 2.37628e-145
N.aligns:None Ident:294 Pos.:294 Gaps:1 Align len:295
Strand: (None, None) Frame: (1, 1)
Query: TCAGCCTACCATGAGAATAAGAGAAAGAAAATGAAGATCAAAAGCTTATTCATCTGTTTTTCTTTTTCGTTGGTGTAAAGCCAACACCCTGTCTAAAAAACATAAATTTCTTTAATCATTTTGCCTCTTTTCTCTGTGCTTCAATTAATAAAAAATGGAAAGAATCTAATAGAGTGGTACAGCACTGTTATTTTTCAAAGATGTGTTGCTATCCTGAAAATTCTGTAGGTTCTGTGGAAGTTCCAGTGTTCTCTCTTATTCCACTTCGGTAGAGGATTTCTAGTTTC-TGTGGGC start: 1883 end: 2176
Match: ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||
Subjc: TCAGCCTACCATGAGAATAAGAGAAAGAAAATGAAGATCAAAAGCTTATTCATCTGTTTTTCTTTTTCGTTGGTGTAAAGCCAACACCCTGTCTAAAAAACATAAATTTCTTTAATCATTTTGCCTCTTTTCTCTGTGCTTCAATTAATAAAAAATGGAAAGAATCTAATAGAGTGGTACAGCACTGTTATTTTTCAAAGATGTGTTGCTATCCTGAAAATTCTGTAGGTTCTGTGGAAGTTCCAGTGTTCTCTCTTATTCCACTTCGGTAGAGGATTTCTAGTTTCTTGTGGGC start: 21834 end: 22128
Score: 449.0 Bits: 406.143 E-val: 1.60396e-109
N.aligns:None Ident:226 Pos.:226 Gaps:0 Align len:227
Strand: (None, None) Frame: (1, 1)
Query: TATCTATCCGTGGTCCTGAACCAGTTATGTGTGTTGCATGAGAAAACGCCAGTAAGTGACAGAGTCACCAAATGCTGCACAGAATCCTTGGTGAACAGGCGACCATGCTTTTCAGCTCTGGAAGTCGATGAAACATACGTTCCCAAAGAGTTTAATGCTGAAACATTCACCTTCCATGCAGATATATGCACACTTTCTGAGAAGGAGAGACAAATCAAGAAACAAAC start: 1459 end: 1685
Match: || ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Subjc: TAGCTATCCGTGGTCCTGAACCAGTTATGTGTGTTGCATGAGAAAACGCCAGTAAGTGACAGAGTCACCAAATGCTGCACAGAATCCTTGGTGAACAGGCGACCATGCTTTTCAGCTCTGGAAGTCGATGAAACATACGTTCCCAAAGAGTTTAATGCTGAAACATTCACCTTCCATGCAGATATATGCACACTTTCTGAGAAGGAGAGACAAATCAAGAAACAAAC start: 18831 end: 19057
Score: 436.0 Bits: 394.421 E-val: 2.90002e-106
N.aligns:None Ident:218 Pos.:218 Gaps:0 Align len:218
Strand: (None, None) Frame: (1, 1)
Query: GGCGGACCTTGCCAAGTATATCTGTGAAAATCAAGATTCGATCTCCAGTAAACTGAAGGAATGCTGTGAAAAACCTCTGTTGGAAAAATCCCACTGCATTGCCGAAGTGGAAAATGATGAGATGCCTGCTGACTTGCCTTCATTAGCTGCTGATTTTGTTGAAAGTAAGGATGTTTGCAAAAACTATGCTGAGGCAAAGGATGTCTTCCTGGGCATGT start: 876 end: 1093
Match: ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Subjc: GGCGGACCTTGCCAAGTATATCTGTGAAAATCAAGATTCGATCTCCAGTAAACTGAAGGAATGCTGTGAAAAACCTCTGTTGGAAAAATCCCACTGCATTGCCGAAGTGGAAAATGATGAGATGCCTGCTGACTTGCCTTCATTAGCTGCTGATTTTGTTGAAAGTAAGGATGTTTGCAAAAACTATGCTGAGGCAAAGGATGTCTTCCTGGGCATGT start: 14165 end: 14382
Score: 425.0 Bits: 384.502 E-val: 5.24337e-103
N.aligns:None Ident:214 Pos.:214 Gaps:0 Align len:215
Strand: (None, None) Frame: (1, 1)
Query: CATACCCTTTTTGGAGACAAATTATGCACAGTTGCAACTCTTCGTGAAACCTATGGTGAAATGGCTGACTGCTGTGCAAAACAAGAACCTGAGAGAAATGAATGCTTCTTGCAACACAAAGATGACAACCCAAACCTCCCCCGATTGGTGAGACCAGAGGTTGATGTGATGTGCACTGCTTTTCATGACAATGAAGAGACATTTTTGAAAAAATA start: 304 end: 518
Match: |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| ||
Subjc: CATACCCTTTTTGGAGACAAATTATGCACAGTTGCAACTCTTCGTGAAACCTATGGTGAAATGGCTGACTGCTGTGCAAAACAAGAACCTGAGAGAAATGAATGCTTCTTGCAACACAAAGATGACAACCCAAACCTCCCCCGATTGGTGAGACCAGAGGTTGATGTGATGTGCACTGCTTTTCATGACAATGAAGAGACATTTTTGAAAAAGTA start: 9340 end: 9554
Score: 278.0 Bits: 251.955 E-val: 2.23348e-63
N.aligns:None Ident:139 Pos.:139 Gaps:0 Align len:139
Strand: (None, None) Frame: (1, 1)
Query: GCTATTAGTTCGTTACACCAAGAAAGTACCCCAAGTGTCAACTCCAACTCTTGTAGAGGTCTCAAGAAACCTAGGAAAAGTGGGCAGCAAATGTTGTAAACATCCTGAAGCAAAAAGAATGCCCTGTGCAGAAGACTAT start: 1323 end: 1461
Match: |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Subjc: GCTATTAGTTCGTTACACCAAGAAAGTACCCCAAGTGTCAACTCCAACTCTTGTAGAGGTCTCAAGAAACCTAGGAAAAGTGGGCAGCAAATGTTGTAAACATCCTGAAGCAAAAAGAATGCCCTGTGCAGAAGACTAT start: 18277 end: 18415
Score: 264.0 Bits: 239.331 E-val: 1.40948e-59
N.aligns:None Ident:132 Pos.:132 Gaps:0 Align len:132
Strand: (None, None) Frame: (1, 1)
Query: GTGTTGATTGCCTTTGCTCAGTATCTTCAGCAGTGTCCATTTGAAGATCATGTAAAATTAGTGAATGAAGTAACTGAATTTGCAAAAACATGTGTTGCTGATGAGTCAGCTGAAAATTGTGACAAATCACTT start: 172 end: 303
Match: ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Subjc: GTGTTGATTGCCTTTGCTCAGTATCTTCAGCAGTGTCCATTTGAAGATCATGTAAAATTAGTGAATGAAGTAACTGAATTTGCAAAAACATGTGTTGCTGATGAGTCAGCTGAAAATTGTGACAAATCACTT start: 7376 end: 7507
Score: 264.0 Bits: 239.331 E-val: 1.40948e-59
N.aligns:None Ident:132 Pos.:132 Gaps:0 Align len:132
Strand: (None, None) Frame: (1, 1)
Query: GGGCAGTAGCTCGCCTGAGCCAGAGATTTCCCAAAGCTGAGTTTGCAGAAGTTTCCAAGTTAGTGACAGATCTTACCAAAGTCCACACGGAATGCTGCCATGGAGATCTGCTTGAATGTGCTGATGACAGGG start: 746 end: 877
Match: ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Subjc: GGGCAGTAGCTCGCCTGAGCCAGAGATTTCCCAAAGCTGAGTTTGCAGAAGTTTCCAAGTTAGTGACAGATCTTACCAAAGTCCACACGGAATGCTGCCATGGAGATCTGCTTGAATGTGCTGATGACAGGG start: 12741 end: 12872
Score: 264.0 Bits: 239.331 E-val: 1.40948e-59
N.aligns:None Ident:138 Pos.:138 Gaps:0 Align len:142
Strand: (None, None) Frame: (1, 1)
Query: AAACAAACTGCACTTGTTGAGCTTGTGAAACACAAGCCCAAGGCAACAAAAGAGCAACTGAAAGCTGTTATGGATGATTTCGCAGCTTTTGTAGAGAAGTGCTGCAAGGCTGACGATAAGGAGACCTGCTTTGCCGAGGAGG start: 1678 end: 1819
Match: |||| | ||||||||||||||| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Subjc: AAACTCAGTGCACTTGTTGAGCTCGTGAAACACAAGCCCAAGGCAACAAAAGAGCAACTGAAAGCTGTTATGGATGATTTCGCAGCTTTTGTAGAGAAGTGCTGCAAGGCTGACGATAAGGAGACCTGCTTTGCCGAGGAGG start: 20245 end: 20386
Score: 261.0 Bits: 236.626 E-val: 1.7171e-58
N.aligns:None Ident:132 Pos.:132 Gaps:0 Align len:133
Strand: (None, None) Frame: (1, 1)
Query: GTTTTTGTATGAATATGCAAGAAGGCATCCTGATTACTCTGTCGTGCTGCTGCTGAGACTTGCCAAGACATATAAAACCACTCTAGAGAAGTGCTGTGCCGCTGCAGATCCTCATGAATGCTATGCCAAAGTG start: 1092 end: 1224
Match: ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Subjc: GTTTTTGTATGAATATGCAAGAAGGCATCCTGATTACTCTGTCGTGCTGCTGCTGAGACTTGCCAAGACATATGAAACCACTCTAGAGAAGTGCTGTGCCGCTGCAGATCCTCATGAATGCTATGCCAAAGTG start: 15781 end: 15913
Score: 256.0 Bits: 232.118 E-val: 2.09185e-57
N.aligns:None Ident:131 Pos.:131 Gaps:0 Align len:133
Strand: (None, None) Frame: (1, 1)
Query: ATACTTATATGAAATTGCCAGAAGACATCCTTACTTCTATGCCCCGGAACTCCTTTTCTTTGCTAAAAGGTATAAAGCTGCTTTTACAGAATGTTGCCAAGCTGCTGATAAGGCTGCCTGCCTGTTGCCAAAG start: 516 end: 648
Match: |||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||
Subjc: ATACTTATATGAAATTGCCAGAAGACATCCTTACTTTTATGCCCCGGAACTCCTTTTCTTTGCTAAAAGGTATAAAGCTGCTTTTACAGAATGTTGCCAAGCTGCTGATAAAGCTGCCTGCCTGTTGCCAAAG start: 10101 end: 10233
Score: 221.0 Bits: 200.559 E-val: 1.23639e-47
N.aligns:None Ident:115 Pos.:115 Gaps:0 Align len:118
Strand: (None, None) Frame: (1, 1)
Query: TCTCTTCTGTCAACCCCACGCGCCTTTGGCACAATGAAGTGGGTAACCTTTATTTCCCTTCTTTTTCTCTTTAGCTCGGCTTATTCCAGGGGTGTGTTTCGTCGAGATGCACACAAGA start: 1 end: 118
Match: ||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| ||||
Subjc: TCTCTTCTGTCAACCCCACACGCCTTTGGCACAATGAAGTGGGTAACCTTTATTTCCCTTCTTTTTCTCTTTAGCTCGGCTTATTCCAGGGGTGTGTTTCGTCGAGATGCACGTAAGA start: 5041 end: 5158
Score: 200.0 Bits: 181.623 E-val: 3.3177e-42
N.aligns:None Ident:100 Pos.:100 Gaps:0 Align len:100
Strand: (None, None) Frame: (1, 1)
Query: GTTCGATGAATTTAAACCTCTTGTGGAAGAGCCTCAGAATTTAATCAAACAAAATTGTGAGCTTTTTGAGCAGCTTGGAGAGTACAAATTCCAGAATGCG start: 1224 end: 1323
Match: ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Subjc: GTTCGATGAATTTAAACCTCTTGTGGAAGAGCCTCAGAATTTAATCAAACAAAATTGTGAGCTTTTTGAGCAGCTTGGAGAGTACAAATTCCAGAATGCG start: 17001 end: 17100
Score: 197.0 Bits: 178.918 E-val: 4.04179e-41
N.aligns:None Ident:100 Pos.:100 Gaps:0 Align len:101
Strand: (None, None) Frame: (1, 1)
Query: AGCTCGATGAACTTCGGGATGAAGGGAAGGCTTCGTCTGCCAAACAGAGGCTCAAGTGTGCCAGTCTCCAAAAATTTGGAGAAAGAGCTTTCAAAGCATGG start: 647 end: 747
Match: ||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||
Subjc: AGCTCGATGAACTTCGGGATGAAGGGAAGGCTTCGTCTGCCAAACAGAGACTCAAGTGTGCCAGTCTCCAAAAATTTGGAGAAAGAGCTTTCAAAGCATGG start: 11056 end: 11156
Score: 135.0 Bits: 123.014 E-val: 2.72574e-24
N.aligns:None Ident:69 Pos.:69 Gaps:0 Align len:70
Strand: (None, None) Frame: (1, 1)
Query: AGGGTAAAAAACTTGTTGCTGCAAGTCGAGCTGCCTTAGGCTTATAACATCACATTTAAAAGCATCTCAG start: 1817 end: 1886
Match: ||||||||||||||||||||||||||| ||||||||||||||||||||||||||||||||||||||||||
Subjc: AGGGTAAAAAACTTGTTGCTGCAAGTCAAGCTGCCTTAGGCTTATAACATCACATTTAAAAGCATCTCAG start: 20998 end: 21067
Score: 117.0 Bits: 106.783 E-val: 2.09555e-19
N.aligns:None Ident:60 Pos.:60 Gaps:0 Align len:61
Strand: (None, None) Frame: (1, 1)
Query: CACACAAGAGTGAGGTTGCTCATCGGTTTAAAGATTTGGGAGAAGAAAATTTCAAAGCCTT start: 110 end: 170
Match: || ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Subjc: CAGACAAGAGTGAGGTTGCTCATCGGTTTAAAGATTTGGGAGAAGAAAATTTCAAAGCCTT start: 5859 end: 5919
Align Title:gi|152112963|gb|EF649953.1| Homo sapiens albumin (ALB) gene, complete cds
Align Len: 21070
Score: 581.0 Bits: 525.165 E-val: 2.37628e-145
N.aligns:None Ident:294 Pos.:294 Gaps:1 Align len:295
Strand: (None, None) Frame: (1, 1)
Query: TCAGCCTACCATGAGAATAAGAGAAAGAAAATGAAGATCAAAAGCTTATTCATCTGTTTTTCTTTTTCGTTGGTGTAAAGCCAACACCCTGTCTAAAAAACATAAATTTCTTTAATCATTTTGCCTCTTTTCTCTGTGCTTCAATTAATAAAAAATGGAAAGAATCTAATAGAGTGGTACAGCACTGTTATTTTTCAAAGATGTGTTGCTATCCTGAAAATTCTGTAGGTTCTGTGGAAGTTCCAGTGTTCTCTCTTATTCCACTTCGGTAGAGGATTTCTAGTTTC-TGTGGGC start: 1883 end: 2176
Match: ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||
Subjc: TCAGCCTACCATGAGAATAAGAGAAAGAAAATGAAGATCAAAAGCTTATTCATCTGTTTTTCTTTTTCGTTGGTGTAAAGCCAACACCCTGTCTAAAAAACATAAATTTCTTTAATCATTTTGCCTCTTTTCTCTGTGCTTCAATTAATAAAAAATGGAAAGAATCTAATAGAGTGGTACAGCACTGTTATTTTTCAAAGATGTGTTGCTATCCTGAAAATTCTGTAGGTTCTGTGGAAGTTCCAGTGTTCTCTCTTATTCCACTTCGGTAGAGGATTTCTAGTTTCTTGTGGGC start: 18784 end: 19078
Score: 449.0 Bits: 406.143 E-val: 1.60396e-109
N.aligns:None Ident:226 Pos.:226 Gaps:0 Align len:227
Strand: (None, None) Frame: (1, 1)
Query: TATCTATCCGTGGTCCTGAACCAGTTATGTGTGTTGCATGAGAAAACGCCAGTAAGTGACAGAGTCACCAAATGCTGCACAGAATCCTTGGTGAACAGGCGACCATGCTTTTCAGCTCTGGAAGTCGATGAAACATACGTTCCCAAAGAGTTTAATGCTGAAACATTCACCTTCCATGCAGATATATGCACACTTTCTGAGAAGGAGAGACAAATCAAGAAACAAAC start: 1459 end: 1685
Match: || ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Subjc: TAGCTATCCGTGGTCCTGAACCAGTTATGTGTGTTGCATGAGAAAACGCCAGTAAGTGACAGAGTCACCAAATGCTGCACAGAATCCTTGGTGAACAGGCGACCATGCTTTTCAGCTCTGGAAGTCGATGAAACATACGTTCCCAAAGAGTTTAATGCTGAAACATTCACCTTCCATGCAGATATATGCACACTTTCTGAGAAGGAGAGACAAATCAAGAAACAAAC start: 15781 end: 16007
Score: 436.0 Bits: 394.421 E-val: 2.90002e-106
N.aligns:None Ident:218 Pos.:218 Gaps:0 Align len:218
Strand: (None, None) Frame: (1, 1)
Query: GGCGGACCTTGCCAAGTATATCTGTGAAAATCAAGATTCGATCTCCAGTAAACTGAAGGAATGCTGTGAAAAACCTCTGTTGGAAAAATCCCACTGCATTGCCGAAGTGGAAAATGATGAGATGCCTGCTGACTTGCCTTCATTAGCTGCTGATTTTGTTGAAAGTAAGGATGTTTGCAAAAACTATGCTGAGGCAAAGGATGTCTTCCTGGGCATGT start: 876 end: 1093
Match: ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Subjc: GGCGGACCTTGCCAAGTATATCTGTGAAAATCAAGATTCGATCTCCAGTAAACTGAAGGAATGCTGTGAAAAACCTCTGTTGGAAAAATCCCACTGCATTGCCGAAGTGGAAAATGATGAGATGCCTGCTGACTTGCCTTCATTAGCTGCTGATTTTGTTGAAAGTAAGGATGTTTGCAAAAACTATGCTGAGGCAAAGGATGTCTTCCTGGGCATGT start: 11115 end: 11332
Score: 425.0 Bits: 384.502 E-val: 5.24337e-103
N.aligns:None Ident:214 Pos.:214 Gaps:0 Align len:215
Strand: (None, None) Frame: (1, 1)
Query: CATACCCTTTTTGGAGACAAATTATGCACAGTTGCAACTCTTCGTGAAACCTATGGTGAAATGGCTGACTGCTGTGCAAAACAAGAACCTGAGAGAAATGAATGCTTCTTGCAACACAAAGATGACAACCCAAACCTCCCCCGATTGGTGAGACCAGAGGTTGATGTGATGTGCACTGCTTTTCATGACAATGAAGAGACATTTTTGAAAAAATA start: 304 end: 518
Match: |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| ||
Subjc: CATACCCTTTTTGGAGACAAATTATGCACAGTTGCAACTCTTCGTGAAACCTATGGTGAAATGGCTGACTGCTGTGCAAAACAAGAACCTGAGAGAAATGAATGCTTCTTGCAACACAAAGATGACAACCCAAACCTCCCCCGATTGGTGAGACCAGAGGTTGATGTGATGTGCACTGCTTTTCATGACAATGAAGAGACATTTTTGAAAAAGTA start: 6290 end: 6504
Score: 278.0 Bits: 251.955 E-val: 2.23348e-63
N.aligns:None Ident:139 Pos.:139 Gaps:0 Align len:139
Strand: (None, None) Frame: (1, 1)
Query: GCTATTAGTTCGTTACACCAAGAAAGTACCCCAAGTGTCAACTCCAACTCTTGTAGAGGTCTCAAGAAACCTAGGAAAAGTGGGCAGCAAATGTTGTAAACATCCTGAAGCAAAAAGAATGCCCTGTGCAGAAGACTAT start: 1323 end: 1461
Match: |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Subjc: GCTATTAGTTCGTTACACCAAGAAAGTACCCCAAGTGTCAACTCCAACTCTTGTAGAGGTCTCAAGAAACCTAGGAAAAGTGGGCAGCAAATGTTGTAAACATCCTGAAGCAAAAAGAATGCCCTGTGCAGAAGACTAT start: 15227 end: 15365
Score: 264.0 Bits: 239.331 E-val: 1.40948e-59
N.aligns:None Ident:132 Pos.:132 Gaps:0 Align len:132
Strand: (None, None) Frame: (1, 1)
Query: GTGTTGATTGCCTTTGCTCAGTATCTTCAGCAGTGTCCATTTGAAGATCATGTAAAATTAGTGAATGAAGTAACTGAATTTGCAAAAACATGTGTTGCTGATGAGTCAGCTGAAAATTGTGACAAATCACTT start: 172 end: 303
Match: ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Subjc: GTGTTGATTGCCTTTGCTCAGTATCTTCAGCAGTGTCCATTTGAAGATCATGTAAAATTAGTGAATGAAGTAACTGAATTTGCAAAAACATGTGTTGCTGATGAGTCAGCTGAAAATTGTGACAAATCACTT start: 4326 end: 4457
Score: 264.0 Bits: 239.331 E-val: 1.40948e-59
N.aligns:None Ident:132 Pos.:132 Gaps:0 Align len:132
Strand: (None, None) Frame: (1, 1)
Query: GGGCAGTAGCTCGCCTGAGCCAGAGATTTCCCAAAGCTGAGTTTGCAGAAGTTTCCAAGTTAGTGACAGATCTTACCAAAGTCCACACGGAATGCTGCCATGGAGATCTGCTTGAATGTGCTGATGACAGGG start: 746 end: 877
Match: ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Subjc: GGGCAGTAGCTCGCCTGAGCCAGAGATTTCCCAAAGCTGAGTTTGCAGAAGTTTCCAAGTTAGTGACAGATCTTACCAAAGTCCACACGGAATGCTGCCATGGAGATCTGCTTGAATGTGCTGATGACAGGG start: 9691 end: 9822
Score: 264.0 Bits: 239.331 E-val: 1.40948e-59
N.aligns:None Ident:138 Pos.:138 Gaps:0 Align len:142
Strand: (None, None) Frame: (1, 1)
Query: AAACAAACTGCACTTGTTGAGCTTGTGAAACACAAGCCCAAGGCAACAAAAGAGCAACTGAAAGCTGTTATGGATGATTTCGCAGCTTTTGTAGAGAAGTGCTGCAAGGCTGACGATAAGGAGACCTGCTTTGCCGAGGAGG start: 1678 end: 1819
Match: |||| | ||||||||||||||| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Subjc: AAACTCAGTGCACTTGTTGAGCTCGTGAAACACAAGCCCAAGGCAACAAAAGAGCAACTGAAAGCTGTTATGGATGATTTCGCAGCTTTTGTAGAGAAGTGCTGCAAGGCTGACGATAAGGAGACCTGCTTTGCCGAGGAGG start: 17195 end: 17336
Score: 261.0 Bits: 236.626 E-val: 1.7171e-58
N.aligns:None Ident:132 Pos.:132 Gaps:0 Align len:133
Strand: (None, None) Frame: (1, 1)
Query: GTTTTTGTATGAATATGCAAGAAGGCATCCTGATTACTCTGTCGTGCTGCTGCTGAGACTTGCCAAGACATATAAAACCACTCTAGAGAAGTGCTGTGCCGCTGCAGATCCTCATGAATGCTATGCCAAAGTG start: 1092 end: 1224
Match: ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Subjc: GTTTTTGTATGAATATGCAAGAAGGCATCCTGATTACTCTGTCGTGCTGCTGCTGAGACTTGCCAAGACATATGAAACCACTCTAGAGAAGTGCTGTGCCGCTGCAGATCCTCATGAATGCTATGCCAAAGTG start: 12731 end: 12863
Score: 256.0 Bits: 232.118 E-val: 2.09185e-57
N.aligns:None Ident:131 Pos.:131 Gaps:0 Align len:133
Strand: (None, None) Frame: (1, 1)
Query: ATACTTATATGAAATTGCCAGAAGACATCCTTACTTCTATGCCCCGGAACTCCTTTTCTTTGCTAAAAGGTATAAAGCTGCTTTTACAGAATGTTGCCAAGCTGCTGATAAGGCTGCCTGCCTGTTGCCAAAG start: 516 end: 648
Match: |||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||
Subjc: ATACTTATATGAAATTGCCAGAAGACATCCTTACTTTTATGCCCCGGAACTCCTTTTCTTTGCTAAAAGGTATAAAGCTGCTTTTACAGAATGTTGCCAAGCTGCTGATAAAGCTGCCTGCCTGTTGCCAAAG start: 7051 end: 7183
Score: 221.0 Bits: 200.559 E-val: 1.23639e-47
N.aligns:None Ident:115 Pos.:115 Gaps:0 Align len:118
Strand: (None, None) Frame: (1, 1)
Query: TCTCTTCTGTCAACCCCACGCGCCTTTGGCACAATGAAGTGGGTAACCTTTATTTCCCTTCTTTTTCTCTTTAGCTCGGCTTATTCCAGGGGTGTGTTTCGTCGAGATGCACACAAGA start: 1 end: 118
Match: ||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| ||||
Subjc: TCTCTTCTGTCAACCCCACACGCCTTTGGCACAATGAAGTGGGTAACCTTTATTTCCCTTCTTTTTCTCTTTAGCTCGGCTTATTCCAGGGGTGTGTTTCGTCGAGATGCACGTAAGA start: 1991 end: 2108
Score: 200.0 Bits: 181.623 E-val: 3.3177e-42
N.aligns:None Ident:100 Pos.:100 Gaps:0 Align len:100
Strand: (None, None) Frame: (1, 1)
Query: GTTCGATGAATTTAAACCTCTTGTGGAAGAGCCTCAGAATTTAATCAAACAAAATTGTGAGCTTTTTGAGCAGCTTGGAGAGTACAAATTCCAGAATGCG start: 1224 end: 1323
Match: ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Subjc: GTTCGATGAATTTAAACCTCTTGTGGAAGAGCCTCAGAATTTAATCAAACAAAATTGTGAGCTTTTTGAGCAGCTTGGAGAGTACAAATTCCAGAATGCG start: 13951 end: 14050
Score: 197.0 Bits: 178.918 E-val: 4.04179e-41
N.aligns:None Ident:100 Pos.:100 Gaps:0 Align len:101
Strand: (None, None) Frame: (1, 1)
Query: AGCTCGATGAACTTCGGGATGAAGGGAAGGCTTCGTCTGCCAAACAGAGGCTCAAGTGTGCCAGTCTCCAAAAATTTGGAGAAAGAGCTTTCAAAGCATGG start: 647 end: 747
Match: ||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||
Subjc: AGCTCGATGAACTTCGGGATGAAGGGAAGGCTTCGTCTGCCAAACAGAGACTCAAGTGTGCCAGTCTCCAAAAATTTGGAGAAAGAGCTTTCAAAGCATGG start: 8006 end: 8106
Score: 135.0 Bits: 123.014 E-val: 2.72574e-24
N.aligns:None Ident:69 Pos.:69 Gaps:0 Align len:70
Strand: (None, None) Frame: (1, 1)
Query: AGGGTAAAAAACTTGTTGCTGCAAGTCGAGCTGCCTTAGGCTTATAACATCACATTTAAAAGCATCTCAG start: 1817 end: 1886
Match: ||||||||||||||||||||||||||| ||||||||||||||||||||||||||||||||||||||||||
Subjc: AGGGTAAAAAACTTGTTGCTGCAAGTCAAGCTGCCTTAGGCTTATAACATCACATTTAAAAGCATCTCAG start: 17948 end: 18017
Score: 117.0 Bits: 106.783 E-val: 2.09555e-19
N.aligns:None Ident:60 Pos.:60 Gaps:0 Align len:61
Strand: (None, None) Frame: (1, 1)
Query: CACACAAGAGTGAGGTTGCTCATCGGTTTAAAGATTTGGGAGAAGAAAATTTCAAAGCCTT start: 110 end: 170
Match: || ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Subjc: CAGACAAGAGTGAGGTTGCTCATCGGTTTAAAGATTTGGGAGAAGAAAATTTCAAAGCCTT start: 2809 end: 2869
Align Title:gi|19387810|gb|AC108157.3| Homo sapiens BAC clone RP11-580P21 from 4, complete sequence
Align Len: 167001
Score: 581.0 Bits: 525.165 E-val: 2.37628e-145
N.aligns:None Ident:294 Pos.:294 Gaps:1 Align len:295
Strand: (None, None) Frame: (1, 1)
Query: TCAGCCTACCATGAGAATAAGAGAAAGAAAATGAAGATCAAAAGCTTATTCATCTGTTTTTCTTTTTCGTTGGTGTAAAGCCAACACCCTGTCTAAAAAACATAAATTTCTTTAATCATTTTGCCTCTTTTCTCTGTGCTTCAATTAATAAAAAATGGAAAGAATCTAATAGAGTGGTACAGCACTGTTATTTTTCAAAGATGTGTTGCTATCCTGAAAATTCTGTAGGTTCTGTGGAAGTTCCAGTGTTCTCTCTTATTCCACTTCGGTAGAGGATTTCTAGTTTC-TGTGGGC start: 1883 end: 2176
Match: ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||
Subjc: TCAGCCTACCATGAGAATAAGAGAAAGAAAATGAAGATCAAAAGCTTATTCATCTGTTTTTCTTTTTCGTTGGTGTAAAGCCAACACCCTGTCTAAAAAACATAAATTTCTTTAATCATTTTGCCTCTTTTCTCTGTGCTTCAATTAATAAAAAATGGAAAGAATCTAATAGAGTGGTACAGCACTGTTATTTTTCAAAGATGTGTTGCTATCCTGAAAATTCTGTAGGTTCTGTGGAAGTTCCAGTGTTCTCTCTTATTCCACTTCGGTAGAGGATTTCTAGTTTCTTGTGGGC start: 86936 end: 87230
Score: 449.0 Bits: 406.143 E-val: 1.60396e-109
N.aligns:None Ident:226 Pos.:226 Gaps:0 Align len:227
Strand: (None, None) Frame: (1, 1)
Query: TATCTATCCGTGGTCCTGAACCAGTTATGTGTGTTGCATGAGAAAACGCCAGTAAGTGACAGAGTCACCAAATGCTGCACAGAATCCTTGGTGAACAGGCGACCATGCTTTTCAGCTCTGGAAGTCGATGAAACATACGTTCCCAAAGAGTTTAATGCTGAAACATTCACCTTCCATGCAGATATATGCACACTTTCTGAGAAGGAGAGACAAATCAAGAAACAAAC start: 1459 end: 1685
Match: || ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Subjc: TAGCTATCCGTGGTCCTGAACCAGTTATGTGTGTTGCATGAGAAAACGCCAGTAAGTGACAGAGTCACCAAATGCTGCACAGAATCCTTGGTGAACAGGCGACCATGCTTTTCAGCTCTGGAAGTCGATGAAACATACGTTCCCAAAGAGTTTAATGCTGAAACATTCACCTTCCATGCAGATATATGCACACTTTCTGAGAAGGAGAGACAAATCAAGAAACAAAC start: 83933 end: 84159
Score: 436.0 Bits: 394.421 E-val: 2.90002e-106
N.aligns:None Ident:218 Pos.:218 Gaps:0 Align len:218
Strand: (None, None) Frame: (1, 1)
Query: GGCGGACCTTGCCAAGTATATCTGTGAAAATCAAGATTCGATCTCCAGTAAACTGAAGGAATGCTGTGAAAAACCTCTGTTGGAAAAATCCCACTGCATTGCCGAAGTGGAAAATGATGAGATGCCTGCTGACTTGCCTTCATTAGCTGCTGATTTTGTTGAAAGTAAGGATGTTTGCAAAAACTATGCTGAGGCAAAGGATGTCTTCCTGGGCATGT start: 876 end: 1093
Match: ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Subjc: GGCGGACCTTGCCAAGTATATCTGTGAAAATCAAGATTCGATCTCCAGTAAACTGAAGGAATGCTGTGAAAAACCTCTGTTGGAAAAATCCCACTGCATTGCCGAAGTGGAAAATGATGAGATGCCTGCTGACTTGCCTTCATTAGCTGCTGATTTTGTTGAAAGTAAGGATGTTTGCAAAAACTATGCTGAGGCAAAGGATGTCTTCCTGGGCATGT start: 79267 end: 79484
Score: 425.0 Bits: 384.502 E-val: 5.24337e-103
N.aligns:None Ident:214 Pos.:214 Gaps:0 Align len:215
Strand: (None, None) Frame: (1, 1)
Query: CATACCCTTTTTGGAGACAAATTATGCACAGTTGCAACTCTTCGTGAAACCTATGGTGAAATGGCTGACTGCTGTGCAAAACAAGAACCTGAGAGAAATGAATGCTTCTTGCAACACAAAGATGACAACCCAAACCTCCCCCGATTGGTGAGACCAGAGGTTGATGTGATGTGCACTGCTTTTCATGACAATGAAGAGACATTTTTGAAAAAATA start: 304 end: 518
Match: |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| ||
Subjc: CATACCCTTTTTGGAGACAAATTATGCACAGTTGCAACTCTTCGTGAAACCTATGGTGAAATGGCTGACTGCTGTGCAAAACAAGAACCTGAGAGAAATGAATGCTTCTTGCAACACAAAGATGACAACCCAAACCTCCCCCGATTGGTGAGACCAGAGGTTGATGTGATGTGCACTGCTTTTCATGACAATGAAGAGACATTTTTGAAAAAGTA start: 74442 end: 74656
Score: 278.0 Bits: 251.955 E-val: 2.23348e-63
N.aligns:None Ident:139 Pos.:139 Gaps:0 Align len:139
Strand: (None, None) Frame: (1, 1)
Query: GCTATTAGTTCGTTACACCAAGAAAGTACCCCAAGTGTCAACTCCAACTCTTGTAGAGGTCTCAAGAAACCTAGGAAAAGTGGGCAGCAAATGTTGTAAACATCCTGAAGCAAAAAGAATGCCCTGTGCAGAAGACTAT start: 1323 end: 1461
Match: |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Subjc: GCTATTAGTTCGTTACACCAAGAAAGTACCCCAAGTGTCAACTCCAACTCTTGTAGAGGTCTCAAGAAACCTAGGAAAAGTGGGCAGCAAATGTTGTAAACATCCTGAAGCAAAAAGAATGCCCTGTGCAGAAGACTAT start: 83379 end: 83517
Score: 264.0 Bits: 239.331 E-val: 1.40948e-59
N.aligns:None Ident:132 Pos.:132 Gaps:0 Align len:132
Strand: (None, None) Frame: (1, 1)
Query: GTGTTGATTGCCTTTGCTCAGTATCTTCAGCAGTGTCCATTTGAAGATCATGTAAAATTAGTGAATGAAGTAACTGAATTTGCAAAAACATGTGTTGCTGATGAGTCAGCTGAAAATTGTGACAAATCACTT start: 172 end: 303
Match: ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Subjc: GTGTTGATTGCCTTTGCTCAGTATCTTCAGCAGTGTCCATTTGAAGATCATGTAAAATTAGTGAATGAAGTAACTGAATTTGCAAAAACATGTGTTGCTGATGAGTCAGCTGAAAATTGTGACAAATCACTT start: 72478 end: 72609
Score: 264.0 Bits: 239.331 E-val: 1.40948e-59
N.aligns:None Ident:132 Pos.:132 Gaps:0 Align len:132
Strand: (None, None) Frame: (1, 1)
Query: GGGCAGTAGCTCGCCTGAGCCAGAGATTTCCCAAAGCTGAGTTTGCAGAAGTTTCCAAGTTAGTGACAGATCTTACCAAAGTCCACACGGAATGCTGCCATGGAGATCTGCTTGAATGTGCTGATGACAGGG start: 746 end: 877
Match: ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Subjc: GGGCAGTAGCTCGCCTGAGCCAGAGATTTCCCAAAGCTGAGTTTGCAGAAGTTTCCAAGTTAGTGACAGATCTTACCAAAGTCCACACGGAATGCTGCCATGGAGATCTGCTTGAATGTGCTGATGACAGGG start: 77843 end: 77974
Score: 264.0 Bits: 239.331 E-val: 1.40948e-59
N.aligns:None Ident:138 Pos.:138 Gaps:0 Align len:142
Strand: (None, None) Frame: (1, 1)
Query: AAACAAACTGCACTTGTTGAGCTTGTGAAACACAAGCCCAAGGCAACAAAAGAGCAACTGAAAGCTGTTATGGATGATTTCGCAGCTTTTGTAGAGAAGTGCTGCAAGGCTGACGATAAGGAGACCTGCTTTGCCGAGGAGG start: 1678 end: 1819
Match: |||| | ||||||||||||||| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Subjc: AAACTCAGTGCACTTGTTGAGCTCGTGAAACACAAGCCCAAGGCAACAAAAGAGCAACTGAAAGCTGTTATGGATGATTTCGCAGCTTTTGTAGAGAAGTGCTGCAAGGCTGACGATAAGGAGACCTGCTTTGCCGAGGAGG start: 85347 end: 85488
Score: 261.0 Bits: 236.626 E-val: 1.7171e-58
N.aligns:None Ident:132 Pos.:132 Gaps:0 Align len:133
Strand: (None, None) Frame: (1, 1)
Query: GTTTTTGTATGAATATGCAAGAAGGCATCCTGATTACTCTGTCGTGCTGCTGCTGAGACTTGCCAAGACATATAAAACCACTCTAGAGAAGTGCTGTGCCGCTGCAGATCCTCATGAATGCTATGCCAAAGTG start: 1092 end: 1224
Match: ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Subjc: GTTTTTGTATGAATATGCAAGAAGGCATCCTGATTACTCTGTCGTGCTGCTGCTGAGACTTGCCAAGACATATGAAACCACTCTAGAGAAGTGCTGTGCCGCTGCAGATCCTCATGAATGCTATGCCAAAGTG start: 80883 end: 81015
Score: 256.0 Bits: 232.118 E-val: 2.09185e-57
N.aligns:None Ident:131 Pos.:131 Gaps:0 Align len:133
Strand: (None, None) Frame: (1, 1)
Query: ATACTTATATGAAATTGCCAGAAGACATCCTTACTTCTATGCCCCGGAACTCCTTTTCTTTGCTAAAAGGTATAAAGCTGCTTTTACAGAATGTTGCCAAGCTGCTGATAAGGCTGCCTGCCTGTTGCCAAAG start: 516 end: 648
Match: |||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||
Subjc: ATACTTATATGAAATTGCCAGAAGACATCCTTACTTTTATGCCCCGGAACTCCTTTTCTTTGCTAAAAGGTATAAAGCTGCTTTTACAGAATGTTGCCAAGCTGCTGATAAAGCTGCCTGCCTGTTGCCAAAG start: 75203 end: 75335
Score: 221.0 Bits: 200.559 E-val: 1.23639e-47
N.aligns:None Ident:115 Pos.:115 Gaps:0 Align len:118
Strand: (None, None) Frame: (1, 1)
Query: TCTCTTCTGTCAACCCCACGCGCCTTTGGCACAATGAAGTGGGTAACCTTTATTTCCCTTCTTTTTCTCTTTAGCTCGGCTTATTCCAGGGGTGTGTTTCGTCGAGATGCACACAAGA start: 1 end: 118
Match: ||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| ||||
Subjc: TCTCTTCTGTCAACCCCACACGCCTTTGGCACAATGAAGTGGGTAACCTTTATTTCCCTTCTTTTTCTCTTTAGCTCGGCTTATTCCAGGGGTGTGTTTCGTCGAGATGCACGTAAGA start: 70143 end: 70260
Score: 200.0 Bits: 181.623 E-val: 3.3177e-42
N.aligns:None Ident:100 Pos.:100 Gaps:0 Align len:100
Strand: (None, None) Frame: (1, 1)
Query: GTTCGATGAATTTAAACCTCTTGTGGAAGAGCCTCAGAATTTAATCAAACAAAATTGTGAGCTTTTTGAGCAGCTTGGAGAGTACAAATTCCAGAATGCG start: 1224 end: 1323
Match: ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Subjc: GTTCGATGAATTTAAACCTCTTGTGGAAGAGCCTCAGAATTTAATCAAACAAAATTGTGAGCTTTTTGAGCAGCTTGGAGAGTACAAATTCCAGAATGCG start: 82103 end: 82202
Score: 197.0 Bits: 178.918 E-val: 4.04179e-41
N.aligns:None Ident:100 Pos.:100 Gaps:0 Align len:101
Strand: (None, None) Frame: (1, 1)
Query: AGCTCGATGAACTTCGGGATGAAGGGAAGGCTTCGTCTGCCAAACAGAGGCTCAAGTGTGCCAGTCTCCAAAAATTTGGAGAAAGAGCTTTCAAAGCATGG start: 647 end: 747
Match: ||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||
Subjc: AGCTCGATGAACTTCGGGATGAAGGGAAGGCTTCGTCTGCCAAACAGAGACTCAAGTGTGCCAGTCTCCAAAAATTTGGAGAAAGAGCTTTCAAAGCATGG start: 76158 end: 76258
Score: 135.0 Bits: 123.014 E-val: 2.72574e-24
N.aligns:None Ident:69 Pos.:69 Gaps:0 Align len:70
Strand: (None, None) Frame: (1, 1)
Query: AGGGTAAAAAACTTGTTGCTGCAAGTCGAGCTGCCTTAGGCTTATAACATCACATTTAAAAGCATCTCAG start: 1817 end: 1886
Match: ||||||||||||||||||||||||||| ||||||||||||||||||||||||||||||||||||||||||
Subjc: AGGGTAAAAAACTTGTTGCTGCAAGTCAAGCTGCCTTAGGCTTATAACATCACATTTAAAAGCATCTCAG start: 86100 end: 86169
Score: 117.0 Bits: 106.783 E-val: 2.09555e-19
N.aligns:None Ident:60 Pos.:60 Gaps:0 Align len:61
Strand: (None, None) Frame: (1, 1)
Query: CACACAAGAGTGAGGTTGCTCATCGGTTTAAAGATTTGGGAGAAGAAAATTTCAAAGCCTT start: 110 end: 170
Match: || ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Subjc: CAGACAAGAGTGAGGTTGCTCATCGGTTTAAAGATTTGGGAGAAGAAAATTTCAAAGCCTT start: 70961 end: 71021
Score: 60.0 Bits: 55.3874 E-val: 0.000332357
N.aligns:None Ident:45 Pos.:45 Gaps:0 Align len:55
Strand: (None, None) Frame: (1, 1)
Query: ATATCTGTGAAAATCAAGATTCGATCTCCAGTAAACTGAAGGAATGCTGTGAAAA start: 893 end: 947
Match: |||| ||| ||| |||||||| |||||||| ||| | || || |||||||||||
Subjc: ATATTTGTTCAAAACAAGATTCTATCTCCAGCAAAATCAAAGAGTGCTGTGAAAA start: 157736 end: 157790
Align Title:gi|178343|gb|M12523.1|HUMALBGC Human serum albumin (ALB) gene, complete cds
Align Len: 19002
Score: 581.0 Bits: 525.165 E-val: 2.37628e-145
N.aligns:None Ident:294 Pos.:294 Gaps:1 Align len:295
Strand: (None, None) Frame: (1, 1)
Query: TCAGCCTACCATGAGAATAAGAGAAAGAAAATGAAGATCAAAAGCTTATTCATCTGTTTTTCTTTTTCGTTGGTGTAAAGCCAACACCCTGTCTAAAAAACATAAATTTCTTTAATCATTTTGCCTCTTTTCTCTGTGCTTCAATTAATAAAAAATGGAAAGAATCTAATAGAGTGGTACAGCACTGTTATTTTTCAAAGATGTGTTGCTATCCTGAAAATTCTGTAGGTTCTGTGGAAGTTCCAGTGTTCTCTCTTATTCCACTTCGGTAGAGGATTTCTAGTTTC-TGTGGGC start: 1883 end: 2176
Match: ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||
Subjc: TCAGCCTACCATGAGAATAAGAGAAAGAAAATGAAGATCAAAAGCTTATTCATCTGTTTTTCTTTTTCGTTGGTGTAAAGCCAACACCCTGTCTAAAAAACATAAATTTCTTTAATCATTTTGCCTCTTTTCTCTGTGCTTCAATTAATAAAAAATGGAAAGAATCTAATAGAGTGGTACAGCACTGTTATTTTTCAAAGATGTGTTGCTATCCTGAAAATTCTGTAGGTTCTGTGGAAGTTCCAGTGTTCTCTCTTATTCCACTTCGGTAGAGGATTTCTAGTTTCTTGTGGGC start: 18522 end: 18816
Score: 449.0 Bits: 406.143 E-val: 1.60396e-109
N.aligns:None Ident:226 Pos.:226 Gaps:0 Align len:227
Strand: (None, None) Frame: (1, 1)
Query: TATCTATCCGTGGTCCTGAACCAGTTATGTGTGTTGCATGAGAAAACGCCAGTAAGTGACAGAGTCACCAAATGCTGCACAGAATCCTTGGTGAACAGGCGACCATGCTTTTCAGCTCTGGAAGTCGATGAAACATACGTTCCCAAAGAGTTTAATGCTGAAACATTCACCTTCCATGCAGATATATGCACACTTTCTGAGAAGGAGAGACAAATCAAGAAACAAAC start: 1459 end: 1685
Match: || ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Subjc: TAGCTATCCGTGGTCCTGAACCAGTTATGTGTGTTGCATGAGAAAACGCCAGTAAGTGACAGAGTCACCAAATGCTGCACAGAATCCTTGGTGAACAGGCGACCATGCTTTTCAGCTCTGGAAGTCGATGAAACATACGTTCCCAAAGAGTTTAATGCTGAAACATTCACCTTCCATGCAGATATATGCACACTTTCTGAGAAGGAGAGACAAATCAAGAAACAAAC start: 15531 end: 15757
Score: 436.0 Bits: 394.421 E-val: 2.90002e-106
N.aligns:None Ident:218 Pos.:218 Gaps:0 Align len:218
Strand: (None, None) Frame: (1, 1)
Query: GGCGGACCTTGCCAAGTATATCTGTGAAAATCAAGATTCGATCTCCAGTAAACTGAAGGAATGCTGTGAAAAACCTCTGTTGGAAAAATCCCACTGCATTGCCGAAGTGGAAAATGATGAGATGCCTGCTGACTTGCCTTCATTAGCTGCTGATTTTGTTGAAAGTAAGGATGTTTGCAAAAACTATGCTGAGGCAAAGGATGTCTTCCTGGGCATGT start: 876 end: 1093
Match: ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Subjc: GGCGGACCTTGCCAAGTATATCTGTGAAAATCAAGATTCGATCTCCAGTAAACTGAAGGAATGCTGTGAAAAACCTCTGTTGGAAAAATCCCACTGCATTGCCGAAGTGGAAAATGATGAGATGCCTGCTGACTTGCCTTCATTAGCTGCTGATTTTGTTGAAAGTAAGGATGTTTGCAAAAACTATGCTGAGGCAAAGGATGTCTTCCTGGGCATGT start: 10866 end: 11083
Score: 425.0 Bits: 384.502 E-val: 5.24337e-103
N.aligns:None Ident:214 Pos.:214 Gaps:0 Align len:215
Strand: (None, None) Frame: (1, 1)
Query: CATACCCTTTTTGGAGACAAATTATGCACAGTTGCAACTCTTCGTGAAACCTATGGTGAAATGGCTGACTGCTGTGCAAAACAAGAACCTGAGAGAAATGAATGCTTCTTGCAACACAAAGATGACAACCCAAACCTCCCCCGATTGGTGAGACCAGAGGTTGATGTGATGTGCACTGCTTTTCATGACAATGAAGAGACATTTTTGAAAAAATA start: 304 end: 518
Match: |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| ||
Subjc: CATACCCTTTTTGGAGACAAATTATGCACAGTTGCAACTCTTCGTGAAACCTATGGTGAAATGGCTGACTGCTGTGCAAAACAAGAACCTGAGAGAAATGAATGCTTCTTGCAACACAAAGATGACAACCCAAACCTCCCCCGATTGGTGAGACCAGAGGTTGATGTGATGTGCACTGCTTTTCATGACAATGAAGAGACATTTTTGAAAAAGTA start: 6041 end: 6255
Score: 278.0 Bits: 251.955 E-val: 2.23348e-63
N.aligns:None Ident:139 Pos.:139 Gaps:0 Align len:139
Strand: (None, None) Frame: (1, 1)
Query: GCTATTAGTTCGTTACACCAAGAAAGTACCCCAAGTGTCAACTCCAACTCTTGTAGAGGTCTCAAGAAACCTAGGAAAAGTGGGCAGCAAATGTTGTAAACATCCTGAAGCAAAAAGAATGCCCTGTGCAGAAGACTAT start: 1323 end: 1461
Match: |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Subjc: GCTATTAGTTCGTTACACCAAGAAAGTACCCCAAGTGTCAACTCCAACTCTTGTAGAGGTCTCAAGAAACCTAGGAAAAGTGGGCAGCAAATGTTGTAAACATCCTGAAGCAAAAAGAATGCCCTGTGCAGAAGACTAT start: 14977 end: 15115
Score: 264.0 Bits: 239.331 E-val: 1.40948e-59
N.aligns:None Ident:132 Pos.:132 Gaps:0 Align len:132
Strand: (None, None) Frame: (1, 1)
Query: GTGTTGATTGCCTTTGCTCAGTATCTTCAGCAGTGTCCATTTGAAGATCATGTAAAATTAGTGAATGAAGTAACTGAATTTGCAAAAACATGTGTTGCTGATGAGTCAGCTGAAAATTGTGACAAATCACTT start: 172 end: 303
Match: ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Subjc: GTGTTGATTGCCTTTGCTCAGTATCTTCAGCAGTGTCCATTTGAAGATCATGTAAAATTAGTGAATGAAGTAACTGAATTTGCAAAAACATGTGTTGCTGATGAGTCAGCTGAAAATTGTGACAAATCACTT start: 4077 end: 4208
Score: 264.0 Bits: 239.331 E-val: 1.40948e-59
N.aligns:None Ident:132 Pos.:132 Gaps:0 Align len:132
Strand: (None, None) Frame: (1, 1)
Query: GGGCAGTAGCTCGCCTGAGCCAGAGATTTCCCAAAGCTGAGTTTGCAGAAGTTTCCAAGTTAGTGACAGATCTTACCAAAGTCCACACGGAATGCTGCCATGGAGATCTGCTTGAATGTGCTGATGACAGGG start: 746 end: 877
Match: ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Subjc: GGGCAGTAGCTCGCCTGAGCCAGAGATTTCCCAAAGCTGAGTTTGCAGAAGTTTCCAAGTTAGTGACAGATCTTACCAAAGTCCACACGGAATGCTGCCATGGAGATCTGCTTGAATGTGCTGATGACAGGG start: 9443 end: 9574
Score: 264.0 Bits: 239.331 E-val: 1.40948e-59
N.aligns:None Ident:138 Pos.:138 Gaps:0 Align len:142
Strand: (None, None) Frame: (1, 1)
Query: AAACAAACTGCACTTGTTGAGCTTGTGAAACACAAGCCCAAGGCAACAAAAGAGCAACTGAAAGCTGTTATGGATGATTTCGCAGCTTTTGTAGAGAAGTGCTGCAAGGCTGACGATAAGGAGACCTGCTTTGCCGAGGAGG start: 1678 end: 1819
Match: |||| | ||||||||||||||| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Subjc: AAACTCAGTGCACTTGTTGAGCTCGTGAAACACAAGCCCAAGGCAACAAAAGAGCAACTGAAAGCTGTTATGGATGATTTCGCAGCTTTTGTAGAGAAGTGCTGCAAGGCTGACGATAAGGAGACCTGCTTTGCCGAGGAGG start: 16933 end: 17074
Score: 261.0 Bits: 236.626 E-val: 1.7171e-58
N.aligns:None Ident:132 Pos.:132 Gaps:0 Align len:133
Strand: (None, None) Frame: (1, 1)
Query: GTTTTTGTATGAATATGCAAGAAGGCATCCTGATTACTCTGTCGTGCTGCTGCTGAGACTTGCCAAGACATATAAAACCACTCTAGAGAAGTGCTGTGCCGCTGCAGATCCTCATGAATGCTATGCCAAAGTG start: 1092 end: 1224
Match: ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Subjc: GTTTTTGTATGAATATGCAAGAAGGCATCCTGATTACTCTGTCGTGCTGCTGCTGAGACTTGCCAAGACATATGAAACCACTCTAGAGAAGTGCTGTGCCGCTGCAGATCCTCATGAATGCTATGCCAAAGTG start: 12481 end: 12613
Score: 256.0 Bits: 232.118 E-val: 2.09185e-57
N.aligns:None Ident:131 Pos.:131 Gaps:0 Align len:133
Strand: (None, None) Frame: (1, 1)
Query: ATACTTATATGAAATTGCCAGAAGACATCCTTACTTCTATGCCCCGGAACTCCTTTTCTTTGCTAAAAGGTATAAAGCTGCTTTTACAGAATGTTGCCAAGCTGCTGATAAGGCTGCCTGCCTGTTGCCAAAG start: 516 end: 648
Match: |||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||
Subjc: ATACTTATATGAAATTGCCAGAAGACATCCTTACTTTTATGCCCCGGAACTCCTTTTCTTTGCTAAAAGGTATAAAGCTGCTTTTACAGAATGTTGCCAAGCTGCTGATAAAGCTGCCTGCCTGTTGCCAAAG start: 6802 end: 6934
Score: 221.0 Bits: 200.559 E-val: 1.23639e-47
N.aligns:None Ident:115 Pos.:115 Gaps:0 Align len:118
Strand: (None, None) Frame: (1, 1)
Query: TCTCTTCTGTCAACCCCACGCGCCTTTGGCACAATGAAGTGGGTAACCTTTATTTCCCTTCTTTTTCTCTTTAGCTCGGCTTATTCCAGGGGTGTGTTTCGTCGAGATGCACACAAGA start: 1 end: 118
Match: ||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| ||||
Subjc: TCTCTTCTGTCAACCCCACACGCCTTTGGCACAATGAAGTGGGTAACCTTTATTTCCCTTCTTTTTCTCTTTAGCTCGGCTTATTCCAGGGGTGTGTTTCGTCGAGATGCACGTAAGA start: 1743 end: 1860
Score: 200.0 Bits: 181.623 E-val: 3.3177e-42
N.aligns:None Ident:100 Pos.:100 Gaps:0 Align len:100
Strand: (None, None) Frame: (1, 1)
Query: GTTCGATGAATTTAAACCTCTTGTGGAAGAGCCTCAGAATTTAATCAAACAAAATTGTGAGCTTTTTGAGCAGCTTGGAGAGTACAAATTCCAGAATGCG start: 1224 end: 1323
Match: ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Subjc: GTTCGATGAATTTAAACCTCTTGTGGAAGAGCCTCAGAATTTAATCAAACAAAATTGTGAGCTTTTTGAGCAGCTTGGAGAGTACAAATTCCAGAATGCG start: 13701 end: 13800
Score: 197.0 Bits: 178.918 E-val: 4.04179e-41
N.aligns:None Ident:100 Pos.:100 Gaps:0 Align len:101
Strand: (None, None) Frame: (1, 1)
Query: AGCTCGATGAACTTCGGGATGAAGGGAAGGCTTCGTCTGCCAAACAGAGGCTCAAGTGTGCCAGTCTCCAAAAATTTGGAGAAAGAGCTTTCAAAGCATGG start: 647 end: 747
Match: ||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||
Subjc: AGCTCGATGAACTTCGGGATGAAGGGAAGGCTTCGTCTGCCAAACAGAGACTCAAGTGTGCCAGTCTCCAAAAATTTGGAGAAAGAGCTTTCAAAGCATGG start: 7757 end: 7857
Score: 135.0 Bits: 123.014 E-val: 2.72574e-24
N.aligns:None Ident:69 Pos.:69 Gaps:0 Align len:70
Strand: (None, None) Frame: (1, 1)
Query: AGGGTAAAAAACTTGTTGCTGCAAGTCGAGCTGCCTTAGGCTTATAACATCACATTTAAAAGCATCTCAG start: 1817 end: 1886
Match: ||||||||||||||||||||||||||| ||||||||||||||||||||||||||||||||||||||||||
Subjc: AGGGTAAAAAACTTGTTGCTGCAAGTCAAGCTGCCTTAGGCTTATAACATCACATTTAAAAGCATCTCAG start: 17686 end: 17755
Score: 117.0 Bits: 106.783 E-val: 2.09555e-19
N.aligns:None Ident:60 Pos.:60 Gaps:0 Align len:61
Strand: (None, None) Frame: (1, 1)
Query: CACACAAGAGTGAGGTTGCTCATCGGTTTAAAGATTTGGGAGAAGAAAATTTCAAAGCCTT start: 110 end: 170
Match: || ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Subjc: CAGACAAGAGTGAGGTTGCTCATCGGTTTAAAGATTTGGGAGAAGAAAATTTCAAAGCCTT start: 2561 end: 2621
Align Title:gi|1036032687|gb|AH007061.2| Homo sapiens chromosome 4 serum albumin (ALB) gene, partial cds; and serum albumin-alphafetoprotein intergenic spacer, partial sequence
Align Len: 523
Score: 579.0 Bits: 523.362 E-val: 8.29405e-145
N.aligns:None Ident:293 Pos.:293 Gaps:1 Align len:294
Strand: (None, None) Frame: (1, 1)
Query: TCAGCCTACCATGAGAATAAGAGAAAGAAAATGAAGATCAAAAGCTTATTCATCTGTTTTTCTTTTTCGTTGGTGTAAAGCCAACACCCTGTCTAAAAAACATAAATTTCTTTAATCATTTTGCCTCTTTTCTCTGTGCTTCAATTAATAAAAAATGGAAAGAATCTAATAGAGTGGTACAGCACTGTTATTTTTCAAAGATGTGTTGCTATCCTGAAAATTCTGTAGGTTCTGTGGAAGTTCCAGTGTTCTCTCTTATTCCACTTCGGTAGAGGATTTCTAGTTTCTGTGGGC start: 1883 end: 2176
Match: |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Subjc: TCAGCCTACCATGAGAATAAGAGAAAGAAAATGAAGATCAAAAGCTTATTCATCTGTTTT-CTTTTTCGTTGGTGTAAAGCCAACACCCTGTCTAAAAAACATAAATTTCTTTAATCATTTTGCCTCTTTTCTCTGTGCTTCAATTAATAAAAAATGGAAAGAATCTAATAGAGTGGTACAGCACTGTTATTTTTCAAAGATGTGTTGCTATCCTGAAAATTCTGTAGGTTCTGTGGAAGTTCCAGTGTTCTCTCTTATTCCACTTCGGTAGAGGATTTCTAGTTTCTGTGGGC start: 4 end: 296
Align Title:gi|408407822|gb|AF116616.2| Homo sapiens PRO0998 mRNA, complete cds
Align Len: 1649
Score: 577.0 Bits: 521.558 E-val: 2.89491e-144
N.aligns:None Ident:290 Pos.:290 Gaps:0 Align len:291
Strand: (None, None) Frame: (1, -1)
Query: AGCTTTTGTAGAGAAGTGCTGCAAGGCTGACGATAAGGAGACCTGCTTTGCCGAGGAGGGTAAAAAACTTGTTGCTGCAAGTCGAGCTGCCTTAGGCTTATAACATCACATTTAAAAGCATCTCAGCCTACCATGAGAATAAGAGAAAGAAAATGAAGATCAAAAGCTTATTCATCTGTTTTTCTTTTTCGTTGGTGTAAAGCCAACACCCTGTCTAAAAAACATAAATTTCTTTAATCATTTTGCCTCTTTTCTCTGTGCTTCAATTAATAAAAAATGGAAAGAATCTAA start: 1761 end: 2051
Match: ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Subjc: AGCTTTTGTAGAGAAGTGCTGCAAGGCTGACGATAAGGAGACCTGCTTTGCCGAGGAGGGTAAAAAACTTGTTGCTGCAAGTCAAGCTGCCTTAGGCTTATAACATCACATTTAAAAGCATCTCAGCCTACCATGAGAATAAGAGAAAGAAAATGAAGATCAAAAGCTTATTCATCTGTTTTTCTTTTTCGTTGGTGTAAAGCCAACACCCTGTCTAAAAAACATAAATTTCTTTAATCATTTTGCCTCTTTTCTCTGTGCTTCAATTAATAAAAAATGGAAAGAATCTAA start: 317 end: 27
Align Title:gi|338858118|gb|AH002596.2|SEG_HUMALB Homo sapiens albumin (ALB) gene, partial cds
Align Len: 1863
Score: 573.0 Bits: 517.951 E-val: 3.52672e-143
N.aligns:None Ident:290 Pos.:290 Gaps:1 Align len:291
Strand: (None, None) Frame: (1, 1)
Query: CCTACCATGAGAATAAGAGAAAGAAAATGAAGATCAAAAGCTTATTCATCTGTTTTTCTTTTTCGTTGGTGTAAAGCCAACACCCTGTCTAAAAAACATAAATTTCTTTAATCATTTTGCCTCTTTTCTCTGTGCTTCAATTAATAAAAAATGGAAAGAATCTAATAGAGTGGTACAGCACTGTTATTTTTCAAAGATGTGTTGCTATCCTGAAAATTCTGTAGGTTCTGTGGAAGTTCCAGTGTTCTCTCTTATTCCACTTCGGTAGAGGATTTCTAGTTTC-TGTGGGC start: 1887 end: 2176
Match: ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||
Subjc: CCTACCATGAGAATAAGAGAAAGAAAATGAAGATCAAAAGCTTATTCATCTGTTTTTCTTTTTCGTTGGTGTAAAGCCAACACCCTGTCTAAAAAACATAAATTTCTTTAATCATTTTGCCTCTTTTCTCTGTGCTTCAATTAATAAAAAATGGAAAGAATCTAATAGAGTGGTACAGCACTGTTATTTTTCAAAGATGTGTTGCTATCCTGAAAATTCTGTAGGTTCTGTGGAAGTTCCAGTGTTCTCTCTTATTCCACTTCGGTAGAGGATTTCTAGTTTCTTGTGGGC start: 1387 end: 1677
Score: 221.0 Bits: 200.559 E-val: 1.23639e-47
N.aligns:None Ident:115 Pos.:115 Gaps:0 Align len:118
Strand: (None, None) Frame: (1, 1)
Query: TCTCTTCTGTCAACCCCACGCGCCTTTGGCACAATGAAGTGGGTAACCTTTATTTCCCTTCTTTTTCTCTTTAGCTCGGCTTATTCCAGGGGTGTGTTTCGTCGAGATGCACACAAGA start: 1 end: 118
Match: ||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| ||||
Subjc: TCTCTTCTGTCAACCCCACACGCCTTTGGCACAATGAAGTGGGTAACCTTTATTTCCCTTCTTTTTCTCTTTAGCTCGGCTTATTCCAGGGGTGTGTTTCGTCGAGATGCACGTAAGA start: 1059 end: 1176
Align Title:gi|1122817323|gb|DQ145726.2| Homo sapiens ARNT-interacting protein 2 (AINP2) mRNA, complete cds
Align Len: 1054
Score: 404.0 Bits: 365.567 E-val: 1.40699e-97
N.aligns:None Ident:205 Pos.:205 Gaps:0 Align len:207
Strand: (None, None) Frame: (1, 1)
Query: AGCTGCCTTAGGCTTATAACATCACATTTAAAAGCATCTCAGCCTACCATGAGAATAAGAGAAAGAAAATGAAGATCAAAAGCTTATTCATCTGTTTTTCTTTTTCGTTGGTGTAAAGCCAACACCCTGTCTAAAAAACATAAATTTCTTTAATCATTTTGCCTCTTTTCTCTGTGCTTCAATTAATAAAAAATGGAAAGAATCTAA start: 1845 end: 2051
Match: ||||||||| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Subjc: AGCTGCCTTTGGCTTATAACATCACATTTAAAAGCATCTCAGCCTACCATGAGAATAAGAGAAAGAAAATGAAGATCAAAAGCTTATTCATCTGTTTTTCTTTTTCGTTGGTGTAAAGCCAACACCCTGCCTAAAAAACATAAATTTCTTTAATCATTTTGCCTCTTTTCTCTGTGCTTCAATTAATAAAAAATGGAAAGAATCTAA start: 836 end: 1042
Align Title:gi|32428|emb|X51365.1| Homo sapiens mRNA for albumin (clone pHA19)
Align Len: 117
Score: 232.0 Bits: 210.477 E-val: 6.8383e-51
N.aligns:None Ident:116 Pos.:116 Gaps:0 Align len:116
Strand: (None, None) Frame: (1, 1)
Query: AGCTTATTCATCTGTTTTTCTTTTTCGTTGGTGTAAAGCCAACACCCTGTCTAAAAAACATAAATTTCTTTAATCATTTTGCCTCTTTTCTCTGTGCTTCAATTAATAAAAAATGG start: 1925 end: 2040
Match: ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Subjc: AGCTTATTCATCTGTTTTTCTTTTTCGTTGGTGTAAAGCCAACACCCTGTCTAAAAAACATAAATTTCTTTAATCATTTTGCCTCTTTTCTCTGTGCTTCAATTAATAAAAAATGG start: 2 end: 117
Align Title:gi|1338686512|ref|NG_056261.1| Homo sapiens albumin (ALB) 5' regulatory region (LOC111832671) on chromosome 4
Align Len: 6858
Score: 221.0 Bits: 200.559 E-val: 1.23639e-47
N.aligns:None Ident:115 Pos.:115 Gaps:0 Align len:118
Strand: (None, None) Frame: (1, 1)
Query: TCTCTTCTGTCAACCCCACGCGCCTTTGGCACAATGAAGTGGGTAACCTTTATTTCCCTTCTTTTTCTCTTTAGCTCGGCTTATTCCAGGGGTGTGTTTCGTCGAGATGCACACAAGA start: 1 end: 118
Match: ||||||||||||||||||| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| ||||
Subjc: TCTCTTCTGTCAACCCCACACGCCTTTGGCACAATGAAGTGGGTAACCTTTATTTCCCTTCTTTTTCTCTTTAGCTCGGCTTATTCCAGGGGTGTGTTTCGTCGAGATGCACGTAAGA start: 6711 end: 6828
Align Title:gi|7959920|gb|AF116711.1|AF116711 Homo sapiens PRO2646 mRNA, complete cds
Align Len: 1160
Score: 216.0 Bits: 196.05 E-val: 1.50624e-46
N.aligns:None Ident:108 Pos.:108 Gaps:0 Align len:108
Strand: (None, None) Frame: (1, -1)
Query: GGAAGAGCCTCAGAATTTAATCAAACAAAATTGTGAGCTTTTTGAGCAGCTTGGAGAGTACAAATTCCAGAATGCGCTATTAGTTCGTTACACCAAGAAAGTACCCCA start: 1248 end: 1355
Match: ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Subjc: GGAAGAGCCTCAGAATTTAATCAAACAAAATTGTGAGCTTTTTGAGCAGCTTGGAGAGTACAAATTCCAGAATGCGCTATTAGTTCGTTACACCAAGAAAGTACCCCA start: 274 end: 167
Example: Let’s align the first 100 bases of the first 5 entries of the file contigs82.fasta to the Malus Domestica genome, writing the results to a apple_first5.xml file. Sample output is here: apple_first5.xml. NOTE: this can take several minutes.
In [40]:
from Bio.Blast import NCBIWWW
from Bio import SeqIO
rc = SeqIO.parse("file_samples/contigs82.fasta", format="fasta")
fasta_string = ""
ids2align =[]
for i in range(5):
entry = next(rc)
fasta_string +=">" + entry.id +"\n"+entry.seq +"\n"
ids2align.append(entry.id)
print("Aligning the following {} entries:\n\t{}".format(
len(ids2align),
"\n\t".join(ids2align)
))
result_handle = NCBIWWW.qblast("blastn", "nt",
fasta_string,
entrez_query='"Malus Domestica" [Organism]'
)
with open("file_samples/blast_res_apple.xml","w") as out_f:
out_f.write(result_handle.read())
print("Output written to \"blast_res_apple.xml\"")
Aligning the following 5 entries:
MDC020656.85
MDC001115.177
MDC013284.379
MDC018185.243
MDC018185.241
Output written to "blast_res_apple.xml"
Getting data from NCBI¶
Biopython provides a module (Bio.Entrez) to pull data off resources
like PubMed or GenBank, and other repositories programmatically through
Entrez.
There are some limitations (mostly taken care directly by Biopython) that you should be aware of when you use NCBI’s services (e.g. they recommend not to run more than 3 requests per second on weekdays, …). Check here if you want to learn what these limitations are.
First of all we need to import the Entrez module with
(from Bio import Entrez) and then we can start interacting with
Entrez, then we should specify (optional) an email setting
Entrez.email (this is optional but you get a warning if you do not
specify it – the email would be used to notify the user in case of
excessive usage).
In particular the module (complete info on Entrez module are here) provides, among the others, the following functions:
res_handle = Entrez.einfo(db)returns a summary of the Entez databases as a results handle.dbis an optional parameter specifying the resource of interest;res_handle = Entrez.esearch(db, term,id)returns all the entries indbhaving query matching the termterm. It is also possible to specify anidto get the information relative to that resource id;res_handle = Entrez.efetch(db, id, rettype, retmode)returns full record corresponding to the identifieridfrom the databasedbformatted inrettype(eg. gb, fasta,… complete list) and return moderetmode(eg. text);res_handle = Entrez.esummary(db, id)returns the summary of the entryidfrom the databasedbas a handle;result = Entrez.read(res_handle)reads the information on the XML handleres_handleand stores them in a dictionary, list or string, depending on the case.
Example:
Let’s get a list of all available databases in Entrez as a dictionary. Let’s then get a summary of the entries in ‘sra’.
In [41]:
from Bio import Entrez
import datetime
Entrez.email = "my_email"
handle = Entrez.einfo()
res = Entrez.read(handle)
#print(res)
print("")
print("As a list:")
print(res['DbList'])
res = Entrez.read(Entrez.einfo(db = "sra"))
#uncomment to see all the information captured
#print(res)
#for el in res["DbInfo"].keys():
# print(el)
date = res["DbInfo"]["LastUpdate"]
dt = datetime.datetime.strptime(date, "%Y/%m/%d %H:%M")
print("")
print("Entries count: {:,}".format(int(res["DbInfo"]["Count"])))
print("LastUpdate: {}/{}/{} {}:{}".format(dt.day,
dt.month,
dt.year,
dt.hour,
dt.minute))
print("Description:", res["DbInfo"]["Description"])
As a list:
['pubmed', 'protein', 'nuccore', 'ipg', 'nucleotide', 'structure', 'sparcle', 'genome', 'annotinfo', 'assembly', 'bioproject', 'biosample', 'blastdbinfo', 'books', 'cdd', 'clinvar', 'gap', 'gapplus', 'grasp', 'dbvar', 'gene', 'gds', 'geoprofiles', 'homologene', 'medgen', 'mesh', 'ncbisearch', 'nlmcatalog', 'omim', 'orgtrack', 'pmc', 'popset', 'probe', 'proteinclusters', 'pcassay', 'biosystems', 'pccompound', 'pcsubstance', 'seqannot', 'snp', 'sra', 'taxonomy', 'biocollections', 'gtr']
Entries count: 9,274,756
LastUpdate: 28/10/2019 11:54
Description: SRA Database
Note that effectively einfo returned a handler to an object that can be read by the read function that produces a dictionary. This dictionary had one key only “DbList” that is the list of available databases in the first case, while the key when db was specified is “DbInfo”.
Example: Fetch the first three entries (retmax = 3) in pubmed that are related to the species “Malus Domestica” and report the title of the publication.
In [42]:
from Bio import Entrez
Entrez.email = "my_email"
handle = Entrez.esearch(db="pubmed", term="Malus Domestica", retmax = 3)
res = Entrez.read(handle)
for el in res.keys():
print(el , " : ", res[el])
print("")
for ids in res["IdList"]:
print("Results for id:", ids)
handle = Entrez.esummary(db="pubmed", id = ids)
res = Entrez.read(handle)
#uncomment to see all info
#print(res)
for r in res:
print(r["Title"])
print(r["Source"])
print("")
Count : 5900
RetMax : 3
RetStart : 0
IdList : ['31655474', '31655083', '31649709']
TranslationSet : [DictElement({'From': 'Malus Domestica', 'To': '"malus"[MeSH Terms] OR "malus"[All Fields] OR ("malus"[All Fields] AND "domestica"[All Fields]) OR "malus domestica"[All Fields]'}, attributes={})]
TranslationStack : [DictElement({'Term': '"malus"[MeSH Terms]', 'Field': 'MeSH Terms', 'Count': '5069', 'Explode': 'Y'}, attributes={}), DictElement({'Term': '"malus"[All Fields]', 'Field': 'All Fields', 'Count': '5907', 'Explode': 'N'}, attributes={}), 'OR', DictElement({'Term': '"malus"[All Fields]', 'Field': 'All Fields', 'Count': '5907', 'Explode': 'N'}, attributes={}), DictElement({'Term': '"domestica"[All Fields]', 'Field': 'All Fields', 'Count': '5891', 'Explode': 'N'}, attributes={}), 'AND', 'GROUP', 'OR', DictElement({'Term': '"malus domestica"[All Fields]', 'Field': 'All Fields', 'Count': '832', 'Explode': 'N'}, attributes={}), 'OR', 'GROUP']
QueryTranslation : "malus"[MeSH Terms] OR "malus"[All Fields] OR ("malus"[All Fields] AND "domestica"[All Fields]) OR "malus domestica"[All Fields]
Results for id: 31655474
The role of glucose-6-phosphate dehydrogenase in reactive oxygen species metabolism in apple exocarp induced by acibenzolar-S-methyl.
Food Chem
Results for id: 31655083
Construction of full-length infectious cDNA clones of Apple stem grooving virus using Gibson Assembly method.
Virus Res
Results for id: 31649709
Red to Brown: An Elevated Anthocyanic Response in Apple Drives Ethylene to Advance Maturity and Fruit Flesh Browning.
Front Plant Sci
Example: Retrieve genbank formatted information of the Malus x domestica MYB domain class transcription factor (MYB1) mRNA complete cds (nucleotide database id:HM122614.1). Parse it as a SeqRecord, printing only the sequence (remember previous practical’s SeqIO).
In [43]:
from Bio import Entrez
from Bio import SeqIO
Entrez.email = "my_email"
handle = Entrez.efetch(db="nucleotide",
id = "HM122614.1",
rettype = "gb",
retmode="text")
my_seq = SeqIO.read(handle, format = "genbank")
print(handle.read())
print(my_seq)
print("")
print("SEQUENCE:")
print(my_seq.seq)
ID: HM122614.1
Name: HM122614
Description: Malus x domestica MYB domain class transcription factor (MYB1) mRNA, complete cds
Number of features: 3
/molecule_type=mRNA
/topology=linear
/data_file_division=HTC
/date=15-AUG-2010
/accessions=['HM122614']
/sequence_version=1
/keywords=['HTC']
/source=Malus domestica (apple)
/organism=Malus domestica
/taxonomy=['Eukaryota', 'Viridiplantae', 'Streptophyta', 'Embryophyta', 'Tracheophyta', 'Spermatophyta', 'Magnoliophyta', 'eudicotyledons', 'Gunneridae', 'Pentapetalae', 'rosids', 'fabids', 'Rosales', 'Rosaceae', 'Maloideae', 'Maleae', 'Malus']
/references=[Reference(title='Transcription Factors in Apple', ...), Reference(title='The FruiTFul database; full length cDNAs of apple transcription factors', ...), Reference(title='Direct Submission', ...)]
Seq('TTTGGTCTGCTGGGTAGGTACTCATAAAAACAAACCAACCGAAGCCTCCGAACC...AAA', IUPACAmbiguousDNA())
SEQUENCE:
TTTGGTCTGCTGGGTAGGTACTCATAAAAACAAACCAACCGAAGCCTCCGAACCGACCACCAATGACGGCCCCAAACGGCGCCGTCCCCAAACAAGCCGACGACCGCCCCGGCACCGAGGCCGAGTTGAACGAGGGCGCAGTGCCCAACGGGAAAGTGAGGGGACCGTGGTCGCCCGAGGAGGACGCGGTGCTGAGCCGGCTCGTGAGCAACTTCGGGGCGAGGAATTGGAGCCTGATCGCCCGAGGAATTCCCGGACGGTCTGGGAAGTCGTGCCGGCTGAGGTGGTGTAATCAGCTTGACCCCTGCGTCAAGCGTAAACCCTTTTCTGAGGAAGAAGACCGTATTATAGTTTCAGCACATGCTATCCATGGGAACAAATGGGCAGTAATTGCAAAGCTTCTTCCAGGTAGAACAGATAACGGAATCAAGAACCATTGGAATTCTACACTAAGGCGCAAGTGCTTTGATAAAGGAAGGTTTAATACTGGACCTGGGGAAATGATGGAAGATGACACCTTTGACAGAAAAAATGCATCCTCAGAAGAAACCCTGTCAGTTGGGAATATCAGTTCATTCAAGACTCATGAAGGAAGAGAGGTCTTGATGGAAAATAGACCAAACCAGTTCGATGTAAGAAGTCATGCAAAGGAGGGCTCTGGCGCTGCCGAATCAAAGCACAATTCTACTCTTATTGCCGAGCCAAGAGACCATCCGACTCTCCAGTCCACCATTTGTTGTCCAGTGGCACGTGTTAGTGCATTTAGTGTTTATAACCGTCCAAGTGTTCCAGCAAATGCTTCATCGTTGTCAAGGACAGTCCCAAGTCATGGCCCTTTGGTCCCAATAACTAAACTAGATTTCGGCTTTGACAACTTCCTTGAAGGTGCATGCAATGAGCCTACGGTTCCTCAACGCTGTGGTCACGGTTGCTGTGATCGAATTGAGGGGCATTCTCAAAGCTCGTTGTTGGGGCCTGAGTTCGTTGAGTATGATGAGCCTCTCCCTTTCTCTAGCCACGAATTAATCTCCATTGCTACGGATTTGAACAAGATTGCATGGATTAAGGGTGGCCTTGAGAGTAATGGGATAAGGATGCCAGAGCACGTAGCAAGCCAGAGAGTCTTTCAAGCAGCTGCCACTACCTTGCAAATGGGGTTGCCAGCAAACACTCCGATGAATGACCATATGCGCTTTGAGGAAGGAAGAAACAAGTTGATGGGAATGATGACGGACGTGCTATCAACCCAGGTGCCTAGACAAACTTTTGCAATGCCAACTGAAGTTGAGGGATTGAGCTAGAACTCATGGCTGGTCTTCCTCAGTCTTCCGGGTGTTTCGGTTAATTTACAGCAGATTAAGTTGAAGTCTCCAGTTTCATTTAGAAGGCTTTTCACTTTTGACTTATCTCGTTTAAGAGGGCATTACATCAAAGATATATATGTAGGTACTTCTCTACCCCTTTTGTCATGAGCAAAGAAGCTGTTGACTAGTAAATGCTTTTATACAAGTGCTTTTATCTCAAATCCGAAAACATAATTGATAAAAAAAAAAAA
Getting data from ExPASy¶
Similarly to what done with Entrez, it is possible to pull data out of ExPASy (https://www.expasy.org/) through the Bio.ExPASy module. We will not cover this in detail. All information can be found here: Bio.ExPASy module.
As an example, we will see how to download a couple of SwissProt entries (the human, rhesus macaque and mouse P53 protein) and perform pairwise sequence alignment of Human vs Macaque and Human vs Mouse.
In [44]:
from Bio import ExPASy
from Bio import SwissProt
from Bio import pairwise2
#the ids of the human, rhesus and mouse proteins
accessions = ["P04637", "P56424", "P02340"]
sequences = []
for accession in accessions:
handle = ExPASy.get_sprot_raw(accession)
record = SwissProt.read(handle)
print("Organism: {}".format(record.organism))
print(record.entry_name)
print(",".join(record.accessions))
print(record.keywords)
print(record.sequence[:30] + "...")
sequences.append(record.sequence)
print("\n\nPairwise sequence alignment of Human vs Rhesus P53:")
aligns = pairwise2.align.globalxx(sequences[0],sequences[1])
for al in aligns[0:3]:
print("Score {}:".format(al[2]))
print(al[0][0:50])
print(al[1][0:50])
print("\n\nPairwise sequence alignment of Human vs Mouse P53:")
aligns = pairwise2.align.globalxx(sequences[0],sequences[2])
for al in aligns[0:3]:
print("Score {}:".format(al[2]))
print(al[0][0:50])
print(al[1][0:50])
Organism: Homo sapiens (Human).
P53_HUMAN
P04637,Q15086,Q15087,Q15088,Q16535,Q16807,Q16808,Q16809,Q16810,Q16811,Q16848,Q2XN98,Q3LRW1,Q3LRW2,Q3LRW3,Q3LRW4,Q3LRW5,Q86UG1,Q8J016,Q99659,Q9BTM4,Q9HAQ8,Q9NP68,Q9NPJ2,Q9NZD0,Q9UBI2,Q9UQ61
['3D-structure', 'Acetylation', 'Activator', 'Alternative promoter usage', 'Alternative splicing', 'Apoptosis', 'Biological rhythms', 'Cell cycle', 'Complete proteome', 'Cytoplasm', 'Direct protein sequencing', 'Disease mutation', 'DNA-binding', 'Endoplasmic reticulum', 'Glycoprotein', 'Host-virus interaction', 'Isopeptide bond', 'Li-Fraumeni syndrome', 'Metal-binding', 'Methylation', 'Mitochondrion', 'Necrosis', 'Nucleus', 'Phosphoprotein', 'Polymorphism', 'Reference proteome', 'Repressor', 'Transcription', 'Transcription regulation', 'Tumor suppressor', 'Ubl conjugation', 'Zinc']
MEEPQSDPSVEPPLSQETFSDLWKLLPENN...
Organism: Macaca mulatta (Rhesus macaque).
P53_MACMU
P56424
['Acetylation', 'Activator', 'Apoptosis', 'Biological rhythms', 'Cell cycle', 'Complete proteome', 'Cytoplasm', 'DNA-binding', 'Endoplasmic reticulum', 'Isopeptide bond', 'Metal-binding', 'Methylation', 'Mitochondrion', 'Necrosis', 'Nucleus', 'Phosphoprotein', 'Reference proteome', 'Repressor', 'Transcription', 'Transcription regulation', 'Tumor suppressor', 'Ubl conjugation', 'Zinc']
MEEPQSDPSIEPPLSQETFSDLWKLLPENN...
Organism: Mus musculus (Mouse).
P53_MOUSE
P02340,Q9QUP3
['3D-structure', 'Acetylation', 'Activator', 'Apoptosis', 'Biological rhythms', 'Cell cycle', 'Complete proteome', 'Cytoplasm', 'Disease mutation', 'DNA-binding', 'Endoplasmic reticulum', 'Isopeptide bond', 'Metal-binding', 'Methylation', 'Mitochondrion', 'Necrosis', 'Nucleus', 'Phosphoprotein', 'Reference proteome', 'Repressor', 'Transcription', 'Transcription regulation', 'Tumor suppressor', 'Ubl conjugation', 'Zinc']
MTAMEESQSDISLELPLSQETFSGLWKLLP...
Pairwise sequence alignment of Human vs Rhesus P53:
Score 376.0:
MEEPQSDPSV-EPPLSQETFSDLWKLLPENNVLSPLPSQAM-DDLMLSPD
MEEPQSDPS-IEPPLSQETFSDLWKLLPENNVLSPLPSQA-VDDLMLSPD
Score 376.0:
MEEPQSDPSVEPPLSQETFSDLWKLLPENNVLSPLPSQAM-DDLMLSPDD
MEEPQSDPSIEPPLSQETFSDLWKLLPENNVLSPLPSQA-VDDLMLSPDD
Score 376.0:
MEEPQSDPSV-EPPLSQETFSDLWKLLPENNVLSPLPSQAMDDLMLSPDD
MEEPQSDPS-IEPPLSQETFSDLWKLLPENNVLSPLPSQAVDDLMLSPDD
Pairwise sequence alignment of Human vs Mouse P53:
Score 315.0:
M---EEP-QSDP-SV-EP-PLSQETFSD-LWKLLPENNVLSP---LPSQA
MTAMEE-SQSD-IS-LE-LPLSQETFS-GLWKLLP------PEDILPS--
Score 315.0:
---MEEP-QSDP-SV-EP-PLSQETFSD-LWKLLPENNVLSP---LPSQA
MTAMEE-SQSD-IS-LE-LPLSQETFS-GLWKLLP------PEDILPS--
Score 315.0:
M---EEPQSDP-SV-EP-PLSQETFSD-LWKLLPENNVLSP---LPSQA-
MTAMEESQSD-IS-LE-LPLSQETFS-GLWKLLP------PEDILPS--P
You can find here all the details on how to deal with the SwissProt records.
3D structure and PDB¶
Biopython can also deal with data coming from the Protein Data Bank database. It is a database of structural information of 3D shapes of proteins, nucleic acids, and complex assemblies. The database currently contains more than 157,000 total structures.
To deal with this kind of data we first need to import Biopython’s
module Bio.PDB with from Bio.PDB import *. All the information
on this module can be found
here.
It is possible to download a structure directly from PDB by using a
PDBList object that features a function called
download_pdb_files having the basic syntax:
PDBList.download_pdb_files(pdb_codes, pdir, file_format)
that downloads the file_format formatted structures defined in the
pdb_codes list of 4 symbols structure Ids from PDB, stores them in
the directory pdir. The safer file_format to use is “mmCif”. The
function will not download the structures more than once. If a file is
already present in the specified directory, a message Structure
exists will be displayed.
Example:
Let’s programmatically download two different structures of the DNA polymerase 3C2K and 3C2L
In [45]:
from Bio.PDB import *
pdbl = PDBList()
structures = ["3C2K", "3C2L"]
el = pdbl.download_pdb_files(structures,
file_format = "mmCif",
pdir = "file_samples/")
Structure exists: 'file_samples/3c2k.cif'
Structure exists: 'file_samples/3c2l.cif'
Macromolecular Crystallographic Information Files (mmCif files .cif) is a paired collection of names (starting with “_”) and values.They also contain a description of the 3D placement of every crystalized atom of the structure. A detailed description of the format can be found here.
Once the structures are available locally, one can start parsing them to
do something useful. Parsing can be done through the MMCIFParser
object:
parser = MMCIFParser()
The parser object has several methods able to deal with structures.
One of these is the get_structure that creates a
PDB.Structure.Structure object with all the data present in the
structure file.
The basic syntax is:
structure = parser.get_structure(pdb_code, filename)
where pdb_code is the PDB code of the structure contained in the
file filename. The method returns a PDB.Structure.Structure that
contains one or more models.
A Structure consists of a collection of one or more Model
(different 3D conformations of the very same structure) that is a
collection of Chain that is a collection of Residues that is a
collection of Atoms. Look in the documentation to get the
information on each of these classes. This is the diagram of a
structure:
Given a Structure we can obtain iterators to models, chains,
residues or atoms with:
Structure.get_models()
Structure.get_chains()
Structure.get_residues()
Structure.get_atoms()
For each model obtained with structure.get_models() function we can
loop through its chains, residues and atoms. For atoms we can get the 3D
coordinates with Atom.get_coord().
Example: Let’s loop through all the models, chain, residues and atoms of the DNA polymerase structure 3C2K. Print the 3D coordinates of each atom.
In [46]:
from Bio.PDB import *
parser = MMCIFParser(QUIET=True) #To disable warnings
filename = "file_samples/3c2l.cif"
structure = parser.get_structure("3c2l", filename)
for model in structure.get_models():
print("model", model, "has {} chains".format(len(model)))
for chain in model:
print(" - chain ", chain, "has {} residues".format(len(chain)))
for residue in chain:
print (" - residue", residue.get_resname(), "has {} atoms".format(len(residue)))
for atom in residue:
x,y,z = atom.get_coord()
print(" - atom:", atom.get_name(), "x: {} y:{} z:{}".format(x,y,z))
model <Model id=0> has 4 chains
- chain <Chain id=T> has 41 residues
- residue DC has 16 atoms
- atom: O5' x: 30.740999221801758 y:-2.2209999561309814 z:16.618999481201172
- atom: C5' x: 31.167999267578125 y:-0.9599999785423279 z:16.062999725341797
- atom: C4' x: 29.996000289916992 y:-0.009999999776482582 z:15.932999610900879
- atom: O4' x: 28.96299934387207 y:-0.6069999933242798 z:15.107000350952148
- atom: C3' x: 29.320999145507812 y:0.38499999046325684 z:17.253000259399414
- atom: O3' x: 29.166000366210938 y:1.8140000104904175 z:17.327999114990234
- atom: C2' x: 27.96500015258789 y:-0.3019999861717224 z:17.187999725341797
- atom: C1' x: 27.701000213623047 y:-0.3490000069141388 z:15.692000389099121
- atom: N1 x: 26.76099967956543 y:-1.4010000228881836 z:15.24899959564209
- atom: C2 x: 25.65999984741211 y:-1.034000039100647 z:14.456999778747559
- atom: O2 x: 25.51300048828125 y:0.16200000047683716 z:14.140999794006348
- atom: N3 x: 24.78499984741211 y:-1.9910000562667847 z:14.057000160217285
- atom: C4 x: 24.97599983215332 y:-3.2660000324249268 z:14.414999961853027
- atom: N4 x: 24.086000442504883 y:-4.172999858856201 z:14.0
- atom: C5 x: 26.08799934387207 y:-3.6679999828338623 z:15.21399974822998
- atom: C6 x: 26.94700050354004 y:-2.7139999866485596 z:15.604000091552734
- residue DC has 19 atoms
- atom: P x: 28.641000747680664 y:2.5 z:18.69099998474121
- atom: OP1 x: 29.559999465942383 y:3.625 z:19.025999069213867
- atom: OP2 x: 28.385000228881836 y:1.4290000200271606 z:19.698999404907227
- atom: O5' x: 27.225000381469727 y:3.124000072479248 z:18.30299949645996
- atom: C5' x: 27.128999710083008 y:4.366000175476074 z:17.600000381469727
- atom: C4' x: 25.67799949645996 y:4.741000175476074 z:17.399999618530273
- atom: O4' x: 24.954999923706055 y:3.5789999961853027 z:16.92799949645996
- atom: C3' x: 24.93600082397461 y:5.171000003814697 z:18.663000106811523
- atom: O3' x: 23.81800079345703 y:5.980000019073486 z:18.29800033569336
- atom: C2' x: 24.413000106811523 y:3.8489999771118164 z:19.187999725341797
- atom: C1' x: 23.961999893188477 y:3.2190001010894775 z:17.882999420166016
- atom: N1 x: 23.832000732421875 y:1.7519999742507935 z:17.87299919128418
- atom: C2 x: 22.68899917602539 y:1.1799999475479126 z:17.298999786376953
- atom: O2 x: 21.812999725341797 y:1.9190000295639038 z:16.829999923706055
- atom: N3 x: 22.569000244140625 y:-0.16200000047683716 z:17.268999099731445
- atom: C4 x: 23.52899932861328 y:-0.9279999732971191 z:17.78700065612793
- atom: N4 x: 23.367000579833984 y:-2.252000093460083 z:17.738000869750977
- atom: C5 x: 24.697999954223633 y:-0.37400001287460327 z:18.3799991607666
- atom: C6 x: 24.808000564575195 y:0.9559999704360962 z:18.40399932861328
- residue DG has 22 atoms
- atom: P x: 23.493000030517578 y:7.309000015258789 z:19.131000518798828
- atom: OP1 x: 24.016000747680664 y:8.472999572753906 z:18.36199951171875
- atom: OP2 x: 23.934999465942383 y:7.0960001945495605 z:20.5310001373291
- atom: O5' x: 21.90399932861328 y:7.382999897003174 z:19.136999130249023
- atom: C5' x: 21.174999237060547 y:7.506999969482422 z:17.922000885009766
- atom: C4' x: 19.93899917602539 y:6.644999980926514 z:17.98200035095215
- atom: O4' x: 20.363000869750977 y:5.269000053405762 z:18.142000198364258
- atom: C3' x: 19.0310001373291 y:6.951000213623047 z:19.179000854492188
- atom: O3' x: 17.66200065612793 y:6.968999862670898 z:18.77199935913086
- atom: C2' x: 19.263999938964844 y:5.7769999504089355 z:20.11400032043457
- atom: C1' x: 19.552000045776367 y:4.665999889373779 z:19.12700080871582
- atom: N9 x: 20.26799964904785 y:3.5169999599456787 z:19.67300033569336
- atom: C8 x: 21.375 y:3.5269999504089355 z:20.490999221801758
- atom: N7 x: 21.760000228881836 y:2.3299999237060547 z:20.850000381469727
- atom: C5 x: 20.85700035095215 y:1.4789999723434448 z:20.225000381469727
- atom: C6 x: 20.763999938964844 y:0.06800000369548798 z:20.246999740600586
- atom: O6 x: 21.47800064086914 y:-0.7459999918937683 z:20.850000381469727
- atom: N1 x: 19.707000732421875 y:-0.382999986410141 z:19.465999603271484
- atom: C2 x: 18.85099983215332 y:0.4169999957084656 z:18.756000518798828
- atom: N2 x: 17.89900016784668 y:-0.20800000429153442 z:18.055999755859375
- atom: N3 x: 18.92099952697754 y:1.7330000400543213 z:18.733999252319336
- atom: C4 x: 19.940000534057617 y:2.194999933242798 z:19.485000610351562
- residue DA has 21 atoms
- atom: P x: 16.55500030517578 y:7.6579999923706055 z:19.7189998626709
- atom: OP1 x: 16.44099998474121 y:9.088000297546387 z:19.329999923706055
- atom: OP2 x: 16.868000030517578 y:7.307000160217285 z:21.135000228881836
- atom: O5' x: 15.189000129699707 y:6.940000057220459 z:19.316999435424805
- atom: C5' x: 14.980999946594238 y:6.421999931335449 z:18.000999450683594
- atom: C4' x: 13.972000122070312 y:5.296999931335449 z:18.041000366210938
- atom: O4' x: 14.60099983215332 y:4.105000019073486 z:18.56999969482422
- atom: C3' x: 12.777000427246094 y:5.566999912261963 z:18.947999954223633
- atom: O3' x: 11.604000091552734 y:4.940000057220459 z:18.440000534057617
- atom: C2' x: 13.196000099182129 y:4.954999923706055 z:20.273000717163086
- atom: C1' x: 14.07699966430664 y:3.7829999923706055 z:19.85700035095215
- atom: N9 x: 15.21399974822998 y:3.496999979019165 z:20.743000030517578
- atom: C8 x: 15.923999786376953 y:4.367000102996826 z:21.542999267578125
- atom: N7 x: 16.92300033569336 y:3.805999994277954 z:22.18000030517578
- atom: C5 x: 16.867000579833984 y:2.4760000705718994 z:21.781999588012695
- atom: C6 x: 17.660999298095703 y:1.3539999723434448 z:22.099000930786133
- atom: N6 x: 18.72599983215332 y:1.3949999809265137 z:22.900999069213867
- atom: N1 x: 17.31800079345703 y:0.1720000058412552 z:21.548999786376953
- atom: C2 x: 16.260000228881836 y:0.12600000202655792 z:20.73200035095215
- atom: N3 x: 15.449000358581543 y:1.1030000448226929 z:20.347999572753906
- atom: C4 x: 15.809000015258789 y:2.2679998874664307 z:20.913000106811523
- residue DC has 19 atoms
- atom: P x: 10.178999900817871 y:5.289999961853027 z:19.089000701904297
- atom: OP1 x: 9.128999710083008 y:5.105999946594238 z:18.054000854492188
- atom: OP2 x: 10.288999557495117 y:6.593999862670898 z:19.79800033569336
- atom: O5' x: 9.972999572753906 y:4.1579999923706055 z:20.18600082397461
- atom: C5' x: 9.642000198364258 y:2.8340001106262207 z:19.795000076293945
- atom: C4' x: 9.98900032043457 y:1.8619999885559082 z:20.893999099731445
- atom: O4' x: 11.369999885559082 y:1.9759999513626099 z:21.277000427246094
- atom: C3' x: 9.21500015258789 y:2.0450000762939453 z:22.19499969482422
- atom: O3' x: 8.050999641418457 y:1.2230000495910645 z:22.155000686645508
- atom: C2' x: 10.194000244140625 y:1.5709999799728394 z:23.26799964904785
- atom: C1' x: 11.472000122070312 y:1.246999979019165 z:22.483999252319336
- atom: N1 x: 12.76099967956543 y:1.5789999961853027 z:23.125999450683594
- atom: C2 x: 13.718000411987305 y:0.5619999766349792 z:23.277999877929688
- atom: O2 x: 13.458999633789062 y:-0.578000009059906 z:22.86400032043457
- atom: N3 x: 14.899999618530273 y:0.8500000238418579 z:23.868999481201172
- atom: C4 x: 15.147000312805176 y:2.0880000591278076 z:24.297000885009766
- atom: N4 x: 16.325000762939453 y:2.3239998817443848 z:24.8700008392334
- atom: C5 x: 14.196999549865723 y:3.1410000324249268 z:24.152000427246094
- atom: C6 x: 13.029999732971191 y:2.8459999561309814 z:23.56599998474121
- residue DC has 19 atoms
- atom: P x: 6.784999847412109 y:1.5880000591278076 z:23.06800079345703
- atom: OP1 x: 7.255000114440918 y:1.5880000591278076 z:24.479000091552734
- atom: OP2 x: 5.660999774932861 y:0.6949999928474426 z:22.66699981689453
- atom: O5' x: 6.434000015258789 y:3.0880000591278076 z:22.659000396728516
- atom: C5' x: 5.370999813079834 y:3.378000020980835 z:21.74799919128418
- atom: C4' x: 4.375999927520752 y:4.328000068664551 z:22.378000259399414
- atom: O4' x: 3.687000036239624 y:3.677000045776367 z:23.465999603271484
- atom: C3' x: 4.9770002365112305 y:5.60699987411499 z:22.972999572753906
- atom: O3' x: 4.706999778747559 y:6.71999979019165 z:22.107999801635742
- atom: C2' x: 4.288000106811523 y:5.76200008392334 z:24.326000213623047
- atom: C1' x: 3.2070000171661377 y:4.689000129699707 z:24.322999954223633
- atom: N1 x: 2.882999897003174 y:4.071000099182129 z:25.628000259399414
- atom: C2 x: 3.9179999828338623 y:3.555999994277954 z:26.45199966430664
- atom: O2 x: 5.10099983215332 y:3.6510000228881836 z:26.08799934387207
- atom: N3 x: 3.5899999141693115 y:2.9709999561309814 z:27.625
- atom: C4 x: 2.311000108718872 y:2.88700008392334 z:27.996999740600586
- atom: N4 x: 2.0390000343322754 y:2.2960000038146973 z:29.16200065612793
- atom: C5 x: 1.2519999742507935 y:3.4059998989105225 z:27.194000244140625
- atom: C6 x: 1.5789999961853027 y:3.9860000610351562 z:26.031999588012695
- residue DG has 22 atoms
- atom: P x: 5.339000225067139 y:8.164999961853027 z:22.430999755859375
- atom: OP1 x: 5.185999870300293 y:9.013999938964844 z:21.215999603271484
- atom: OP2 x: 6.695000171661377 y:7.960000038146973 z:23.01799964904785
- atom: O5' x: 4.388999938964844 y:8.75 z:23.570999145507812
- atom: C5' x: 2.9639999866485596 y:8.717000007629395 z:23.433000564575195
- atom: C4' x: 2.2890000343322754 y:8.987000465393066 z:24.761999130249023
- atom: O4' x: 2.5959999561309814 y:7.940000057220459 z:25.718000411987305
- atom: C3' x: 2.671999931335449 y:10.305999755859375 z:25.44499969482422
- atom: O3' x: 1.5190000534057617 y:10.883999824523926 z:26.075000762939453
- atom: C2' x: 3.6530001163482666 y:9.859000205993652 z:26.516000747680664
- atom: C1' x: 3.0190000534057617 y:8.543000221252441 z:26.93000030517578
- atom: N9 x: 3.871000051498413 y:7.5920000076293945 z:27.639999389648438
- atom: C8 x: 5.236999988555908 y:7.434000015258789 z:27.535999298095703
- atom: N7 x: 5.703999996185303 y:6.49399995803833 z:28.315000534057617
- atom: C5 x: 4.581999778747559 y:5.999000072479248 z:28.972000122070312
- atom: C6 x: 4.456999778747559 y:4.964000225067139 z:29.95199966430664
- atom: O6 x: 5.3429999351501465 y:4.252999782562256 z:30.444000244140625
- atom: N1 x: 3.134000062942505 y:4.795000076293945 z:30.350000381469727
- atom: C2 x: 2.063999891281128 y:5.514999866485596 z:29.8700008392334
- atom: N2 x: 0.8629999756813049 y:5.197000026702881 z:30.371000289916992
- atom: N3 x: 2.1649999618530273 y:6.474999904632568 z:28.964000701904297
- atom: C4 x: 3.443000078201294 y:6.664000034332275 z:28.562999725341797
- residue DC has 19 atoms
- atom: P x: 0.33799999952316284 y:11.519000053405762 z:25.18000030517578
- atom: OP1 x: 0.1550000011920929 y:10.725000381469727 z:23.934999465942383
- atom: OP2 x: 0.6060000061988831 y:12.97599983215332 z:25.08799934387207
- atom: O5' x: -0.9620000123977661 y:11.310999870300293 z:26.07699966430664
- atom: C5' x: -1.5479999780654907 y:10.01099967956543 z:26.243999481201172
- atom: C4' x: -2.0759999752044678 y:9.840999603271484 z:27.652999877929688
- atom: O4' x: -1.0989999771118164 y:9.121000289916992 z:28.450000762939453
- atom: C3' x: -2.3559999465942383 y:11.156999588012695 z:28.393999099731445
- atom: O3' x: -3.6589999198913574 y:11.140999794006348 z:28.993000030517578
- atom: C2' x: -1.2630000114440918 y:11.21500015258789 z:29.448999404907227
- atom: C1' x: -1.0080000162124634 y:9.743000030517578 z:29.716999053955078
- atom: N1 x: 0.3089999854564667 y:9.418000221252441 z:30.30299949645996
- atom: C2 x: 0.3610000014305115 y:8.574000358581543 z:31.42300033569336
- atom: O2 x: -0.6980000138282776 y:8.114999771118164 z:31.885000228881836
- atom: N3 x: 1.565000057220459 y:8.277999877929688 z:31.972000122070312
- atom: C4 x: 2.681999921798706 y:8.782999992370605 z:31.444000244140625
- atom: N4 x: 3.8420000076293945 y:8.456999778747559 z:32.00699996948242
- atom: C5 x: 2.6589999198913574 y:9.640000343322754 z:30.309999465942383
- atom: C6 x: 1.4630000591278076 y:9.928999900817871 z:29.774999618530273
- residue DG has 22 atoms
- atom: P x: -4.367000102996826 y:12.529000282287598 z:29.423999786376953
- atom: OP1 x: -5.24399995803833 y:12.972000122070312 z:28.31399917602539
- atom: OP2 x: -3.3519999980926514 y:13.472999572753906 z:29.975000381469727
- atom: O5' x: -5.328999996185303 y:12.105999946594238 z:30.618999481201172
- atom: C5' x: -5.7230000495910645 y:10.743000030517578 z:30.81599998474121
- atom: C4' x: -5.9730000495910645 y:10.482000350952148 z:32.2859992980957
- atom: O4' x: -4.754000186920166 y:10.02400016784668 z:32.917999267578125
- atom: C3' x: -6.394000053405762 y:11.713000297546387 z:33.08300018310547
- atom: O3' x: -7.164999961853027 y:11.305000305175781 z:34.207000732421875
- atom: C2' x: -5.059999942779541 y:12.281000137329102 z:33.53200149536133
- atom: C1' x: -4.284999847412109 y:11.005999565124512 z:33.8380012512207
- atom: N9 x: -2.8359999656677246 y:11.102999687194824 z:33.691001892089844
- atom: C8 x: -2.1500000953674316 y:11.842000007629395 z:32.762001037597656
- atom: N7 x: -0.8539999723434448 y:11.725000381469727 z:32.86600112915039
- atom: C5 x: -0.6690000295639038 y:10.854999542236328 z:33.930999755859375
- atom: C6 x: 0.5249999761581421 y:10.35200023651123 z:34.5099983215332
- atom: O6 x: 1.6929999589920044 y:10.571999549865723 z:34.17399978637695
- atom: N1 x: 0.25699999928474426 y:9.508999824523926 z:35.584999084472656
- atom: C2 x: -0.9919999837875366 y:9.182999610900879 z:36.04100036621094
- atom: N2 x: -1.0390000343322754 y:8.371000289916992 z:37.106998443603516
- atom: N3 x: -2.114000082015991 y:9.626999855041504 z:35.5
- atom: C4 x: -1.8799999952316284 y:10.461000442504883 z:34.457000732421875
- residue DC has 19 atoms
- atom: P x: -7.857999801635742 y:12.404000282287598 z:35.13100051879883
- atom: OP1 x: -9.21500015258789 y:11.907999992370605 z:35.45399856567383
- atom: OP2 x: -7.696000099182129 y:13.723999977111816 z:34.48099899291992
- atom: O5' x: -6.958000183105469 y:12.425999641418457 z:36.45100021362305
- atom: C5' x: -7.033999919891357 y:11.376999855041504 z:37.417999267578125
- atom: C4' x: -5.870999813079834 y:11.456999778747559 z:38.382999420166016
- atom: O4' x: -4.645999908447266 y:11.569000244140625 z:37.64799880981445
- atom: C3' x: -5.867000102996826 y:12.637999534606934 z:39.358001708984375
- atom: O3' x: -6.331999778747559 y:12.196000099182129 z:40.64400100708008
- atom: C2' x: -4.410999774932861 y:13.088000297546387 z:39.395999908447266
- atom: C1' x: -3.680999994277954 y:12.074999809265137 z:38.53200149536133
- atom: N1 x: -2.5789999961853027 y:12.61400032043457 z:37.72200012207031
- atom: C2 x: -1.2640000581741333 y:12.213000297546387 z:38.012001037597656
- atom: O2 x: -1.0640000104904175 y:11.440999984741211 z:38.959999084472656
- atom: N3 x: -0.24699999392032623 y:12.678999900817871 z:37.25299835205078
- atom: C4 x: -0.4959999918937683 y:13.519000053405762 z:36.24800109863281
- atom: N4 x: 0.5400000214576721 y:13.949999809265137 z:35.527000427246094
- atom: C5 x: -1.8209999799728394 y:13.954999923706055 z:35.93899917602539
- atom: C6 x: -2.822999954223633 y:13.482999801635742 z:36.69499969482422
- residue DA has 21 atoms
- atom: P x: -6.264999866485596 y:13.177000045776367 z:41.92599868774414
- atom: OP1 x: -7.065999984741211 y:12.534000396728516 z:43.00299835205078
- atom: OP2 x: -6.591000080108643 y:14.555000305175781 z:41.50400161743164
- atom: O5' x: -4.739999771118164 y:13.109999656677246 z:42.375999450683594
- atom: C5' x: -4.144000053405762 y:11.85200023651123 z:42.70800018310547
- atom: C4' x: -2.9830000400543213 y:12.027999877929688 z:43.66400146484375
- atom: O4' x: -1.7519999742507935 y:12.300000190734863 z:42.95500183105469
- atom: C3' x: -3.1029999256134033 y:13.125 z:44.70899963378906
- atom: O3' x: -2.431999921798706 y:12.666999816894531 z:45.887001037597656
- atom: C2' x: -2.390000104904175 y:14.291999816894531 z:44.04800033569336
- atom: C1' x: -1.2949999570846558 y:13.611000061035156 z:43.23400115966797
- atom: N9 x: -0.9739999771118164 y:14.23900032043457 z:41.952999114990234
- atom: C8 x: -1.8300000429153442 y:14.854999542236328 z:41.07400131225586
- atom: N7 x: -1.25 y:15.282999992370605 z:39.97800064086914
- atom: C5 x: 0.07800000160932541 y:14.930999755859375 z:40.14799880981445
- atom: C6 x: 1.2029999494552612 y:15.083999633789062 z:39.32500076293945
- atom: N6 x: 1.1610000133514404 y:15.645000457763672 z:38.11600112915039
- atom: N1 x: 2.38700008392334 y:14.62600040435791 z:39.78900146484375
- atom: C2 x: 2.4230000972747803 y:14.048999786376953 z:40.99599838256836
- atom: N3 x: 1.430999994277954 y:13.84000015258789 z:41.85900115966797
- atom: C4 x: 0.26899999380111694 y:14.305000305175781 z:41.36800003051758
- residue DT has 20 atoms
- atom: P x: -2.188999891281128 y:13.661999702453613 z:47.132999420166016
- atom: OP1 x: -2.315999984741211 y:12.843000411987305 z:48.37300109863281
- atom: OP2 x: -2.986999988555908 y:14.913000106811523 z:47.0
- atom: O5' x: -0.6570000052452087 y:14.04800033569336 z:46.965999603271484
- atom: C5' x: 0.3330000042915344 y:13.050999641418457 z:46.792999267578125
- atom: C4' x: 1.6670000553131104 y:13.706999778747559 z:46.54499816894531
- atom: O4' x: 1.6430000066757202 y:14.303999900817871 z:45.2239990234375
- atom: C3' x: 1.9980000257492065 y:14.854999542236328 z:47.505001068115234
- atom: O3' x: 3.4089999198913574 y:14.859000205993652 z:47.75600051879883
- atom: C2' x: 1.628999948501587 y:16.091999053955078 z:46.70199966430664
- atom: C1' x: 2.1050000190734863 y:15.644000053405762 z:45.33000183105469
- atom: N1 x: 1.6460000276565552 y:16.400999069213867 z:44.13199996948242
- atom: C2 x: 2.572999954223633 y:16.636999130249023 z:43.12900161743164
- atom: O2 x: 3.7309999465942383 y:16.2810001373291 z:43.194000244140625
- atom: N3 x: 2.0889999866485596 y:17.312999725341797 z:42.03900146484375
- atom: C4 x: 0.8059999942779541 y:17.77199935913086 z:41.84700012207031
- atom: O4 x: 0.5199999809265137 y:18.349000930786133 z:40.797000885009766
- atom: C5 x: -0.11999999731779099 y:17.506999969482422 z:42.948001861572266
- atom: C7 x: -1.534999966621399 y:17.979000091552734 z:42.840999603271484
- atom: C6 x: 0.3440000116825104 y:16.844999313354492 z:44.018001556396484
- residue DC has 19 atoms
- atom: P x: 3.9679999351501465 y:15.352999687194824 z:49.18199920654297
- atom: OP1 x: 4.440999984741211 y:14.140000343322754 z:49.9010009765625
- atom: OP2 x: 2.9700000286102295 y:16.26300048828125 z:49.823001861572266
- atom: O5' x: 5.254000186920166 y:16.208999633789062 z:48.83000183105469
- atom: C5' x: 6.129000186920166 y:15.824000358581543 z:47.78900146484375
- atom: C4' x: 6.697999954223633 y:17.05900001525879 z:47.137001037597656
- atom: O4' x: 5.788000106811523 y:17.520000457763672 z:46.10200119018555
- atom: C3' x: 6.85699987411499 y:18.238000869750977 z:48.10900115966797
- atom: O3' x: 8.050000190734863 y:18.95199966430664 z:47.797000885009766
- atom: C2' x: 5.665999889373779 y:19.121000289916992 z:47.775001525878906
- atom: C1' x: 5.61899995803833 y:18.917999267578125 z:46.27199935913086
- atom: N1 x: 4.400000095367432 y:19.339000701904297 z:45.55400085449219
- atom: C2 x: 4.5269999504089355 y:19.80699920654297 z:44.236000061035156
- atom: O2 x: 5.656000137329102 y:19.850000381469727 z:43.71200180053711
- atom: N3 x: 3.4189999103546143 y:20.195999145507812 z:43.56399917602539
- atom: C4 x: 2.2239999771118164 y:20.1200008392334 z:44.14899826049805
- atom: N4 x: 1.159999966621399 y:20.499000549316406 z:43.44300079345703
- atom: C5 x: 2.065999984741211 y:19.648000717163086 z:45.48500061035156
- atom: C6 x: 3.1689999103546143 y:19.274999618530273 z:46.145999908447266
- residue DA has 21 atoms
- atom: P x: 9.348999977111816 y:18.784000396728516 z:48.71699905395508
- atom: OP1 x: 9.670999526977539 y:17.33099937438965 z:48.79199981689453
- atom: OP2 x: 9.105999946594238 y:19.55500030517578 z:49.965999603271484
- atom: O5' x: 10.47599983215332 y:19.483999252319336 z:47.83599853515625
- atom: C5' x: 10.871000289916992 y:18.892000198364258 z:46.59400177001953
- atom: C4' x: 11.45199966430664 y:19.92099952697754 z:45.64799880981445
- atom: O4' x: 10.427000045776367 y:20.615999221801758 z:44.90599822998047
- atom: C3' x: 12.335000038146973 y:21.007999420166016 z:46.249000549316406
- atom: O3' x: 13.369999885559082 y:21.326000213623047 z:45.31800079345703
- atom: C2' x: 11.381999969482422 y:22.177000045776367 z:46.426998138427734
- atom: C1' x: 10.312000274658203 y:21.95800018310547 z:45.35599899291992
- atom: N9 x: 8.944000244140625 y:22.131000518798828 z:45.83399963378906
- atom: C8 x: 8.444000244140625 y:21.815000534057617 z:47.073001861572266
- atom: N7 x: 7.1620001792907715 y:22.04400062561035 z:47.196998596191406
- atom: C5 x: 6.795000076293945 y:22.551000595092773 z:45.95899963378906
- atom: C6 x: 5.565000057220459 y:22.979999542236328 z:45.452999114990234
- atom: N6 x: 4.434999942779541 y:22.95800018310547 z:46.16299819946289
- atom: N1 x: 5.5329999923706055 y:23.437000274658203 z:44.18199920654297
- atom: C2 x: 6.673999786376953 y:23.458999633789062 z:43.481998443603516
- atom: N3 x: 7.896999835968018 y:23.07699966430664 z:43.85300064086914
- atom: C4 x: 7.885000228881836 y:22.624000549316406 z:45.11600112915039
- residue DG has 22 atoms
- atom: P x: 14.439000129699707 y:22.465999603271484 z:45.67100143432617
- atom: OP1 x: 15.732000350952148 y:22.033000946044922 z:45.07500076293945
- atom: OP2 x: 14.36400032043457 y:22.801000595092773 z:47.12300109863281
- atom: O5' x: 13.901000022888184 y:23.725000381469727 z:44.86199951171875
- atom: C5' x: 13.479999542236328 y:23.606000900268555 z:43.500999450683594
- atom: C4' x: 12.72700023651123 y:24.849000930786133 z:43.0880012512207
- atom: O4' x: 11.375 y:24.797000885009766 z:43.606998443603516
- atom: C3' x: 13.336999893188477 y:26.138999938964844 z:43.637001037597656
- atom: O3' x: 13.461999893188477 y:27.09600067138672 z:42.59199905395508
- atom: C2' x: 12.329000473022461 y:26.611000061035156 z:44.67499923706055
- atom: C1' x: 11.03499984741211 y:26.066999435424805 z:44.11199951171875
- atom: N9 x: 9.951000213623047 y:25.892000198364258 z:45.073001861572266
- atom: C8 x: 10.029000282287598 y:25.426000595092773 z:46.36800003051758
- atom: N7 x: 8.871000289916992 y:25.405000686645508 z:46.97800064086914
- atom: C5 x: 7.974999904632568 y:25.875999450683594 z:46.02299880981445
- atom: C6 x: 6.565999984741211 y:26.079999923706055 z:46.090999603271484
- atom: O6 x: 5.789000034332275 y:25.85099983215332 z:47.0369987487793
- atom: N1 x: 6.070000171661377 y:26.593000411987305 z:44.902000427246094
- atom: C2 x: 6.811999797821045 y:26.860000610351562 z:43.779998779296875
- atom: N2 x: 6.139999866485596 y:27.356000900268555 z:42.720001220703125
- atom: N3 x: 8.112000465393066 y:26.660999298095703 z:43.69499969482422
- atom: C4 x: 8.626999855041504 y:26.17799949645996 z:44.845001220703125
- residue DC has 19 atoms
- atom: P x: 14.321999549865723 y:28.42799949645996 z:42.8380012512207
- atom: OP1 x: 15.064000129699707 y:28.68000030517578 z:41.57600021362305
- atom: OP2 x: 15.067000389099121 y:28.295000076293945 z:44.112998962402344
- atom: O5' x: 13.229000091552734 y:29.56800079345703 z:43.03499984741211
- atom: C5' x: 12.413000106811523 y:29.974000930786133 z:41.94300079345703
- atom: C4' x: 11.097000122070312 y:30.51300048828125 z:42.446998596191406
- atom: O4' x: 10.517000198364258 y:29.645000457763672 z:43.43600082397461
- atom: C3' x: 11.199999809265137 y:31.851999282836914 z:43.159000396728516
- atom: O3' x: 11.206000328063965 y:32.849998474121094 z:42.15399932861328
- atom: C2' x: 9.986000061035156 y:31.849000930786133 z:44.07699966430664
- atom: C1' x: 9.475000381469727 y:30.405000686645508 z:44.016998291015625
- atom: N1 x: 9.112000465393066 y:29.808000564575195 z:45.314998626708984
- atom: C2 x: 7.763000011444092 y:29.753999710083008 z:45.65299987792969
- atom: O2 x: 6.926000118255615 y:30.17300033569336 z:44.83599853515625
- atom: N3 x: 7.3979997634887695 y:29.2450008392334 z:46.8489990234375
- atom: C4 x: 8.324999809265137 y:28.78700065612793 z:47.6870002746582
- atom: N4 x: 7.909999847412109 y:28.29199981689453 z:48.85599899291992
- atom: C5 x: 9.71399974822998 y:28.812999725341797 z:47.36399841308594
- atom: C6 x: 10.059000015258789 y:29.32699966430664 z:46.17900085449219
- residue MN has 1 atoms
- atom: MN x: 23.54800033569336 y:1.0 z:22.975000381469727
- residue HOH has 1 atoms
- atom: O x: -0.03799999877810478 y:22.62299919128418 z:45.85499954223633
- residue HOH has 1 atoms
- atom: O x: -0.2680000066757202 y:6.394999980926514 z:26.27400016784668
- residue HOH has 1 atoms
- atom: O x: -2.617000102996826 y:17.14299964904785 z:38.38199996948242
- residue HOH has 1 atoms
- atom: O x: -2.8450000286102295 y:9.204000473022461 z:40.37200164794922
- residue HOH has 1 atoms
- atom: O x: -4.395999908447266 y:15.696999549865723 z:41.887001037597656
- residue HOH has 1 atoms
- atom: O x: 8.746000289916992 y:3.684000015258789 z:24.974000930786133
- residue HOH has 1 atoms
- atom: O x: 20.542999267578125 y:3.009999990463257 z:24.375
- residue HOH has 1 atoms
- atom: O x: -6.396999835968018 y:10.779000282287598 z:28.01799964904785
- residue HOH has 1 atoms
- atom: O x: -9.277000427246094 y:15.675000190734863 z:35.47200012207031
- residue HOH has 1 atoms
- atom: O x: 6.900000095367432 y:24.628000259399414 z:49.99599838256836
- residue HOH has 1 atoms
- atom: O x: 30.777999877929688 y:4.184000015258789 z:16.06800079345703
- residue HOH has 1 atoms
- atom: O x: 14.788999557495117 y:20.408000946044922 z:42.99700164794922
- residue HOH has 1 atoms
- atom: O x: -2.247999906539917 y:17.725000381469727 z:46.025001525878906
- residue HOH has 1 atoms
- atom: O x: 25.37700080871582 y:2.634999990463257 z:22.086999893188477
- residue HOH has 1 atoms
- atom: O x: 24.393999099731445 y:-1.1100000143051147 z:22.084999084472656
- residue HOH has 1 atoms
- atom: O x: 5.159999847412109 y:11.8100004196167 z:48.76900100708008
- residue HOH has 1 atoms
- atom: O x: -1.069000005722046 y:4.329999923706055 z:24.06399917602539
- residue HOH has 1 atoms
- atom: O x: -0.26600000262260437 y:7.039999961853027 z:23.948999404907227
- residue HOH has 1 atoms
- atom: O x: 7.377999782562256 y:9.512999534606934 z:17.650999069213867
- residue HOH has 1 atoms
- atom: O x: 9.468000411987305 y:9.222999572753906 z:19.29199981689453
- residue HOH has 1 atoms
- atom: O x: 10.92199993133545 y:19.01799964904785 z:41.494998931884766
- residue HOH has 1 atoms
- atom: O x: -5.491000175476074 y:14.925999641418457 z:35.11899948120117
- residue HOH has 1 atoms
- atom: O x: -8.88599967956543 y:16.402999877929688 z:38.94599914550781
- residue HOH has 1 atoms
- atom: O x: -9.723999977111816 y:17.750999450683594 z:33.974998474121094
- chain <Chain id=P> has 18 residues
- residue DG has 19 atoms
- atom: O5' x: -2.617000102996826 y:29.32900047302246 z:49.946998596191406
- atom: C5' x: -3.3010001182556152 y:30.40399932861328 z:49.26599884033203
- atom: C4' x: -2.434000015258789 y:30.941999435424805 z:48.154998779296875
- atom: O4' x: -1.0850000381469727 y:30.972999572753906 z:48.6510009765625
- atom: C3' x: -2.38700008392334 y:30.049999237060547 z:46.91699981689453
- atom: O3' x: -3.3589999675750732 y:30.52400016784668 z:45.96099853515625
- atom: C2' x: -0.9549999833106995 y:30.20599937438965 z:46.415000915527344
- atom: C1' x: -0.17599999904632568 y:30.812999725341797 z:47.584999084472656
- atom: N9 x: 0.9449999928474426 y:30.017000198364258 z:48.07699966430664
- atom: C8 x: 0.9980000257492065 y:29.2549991607666 z:49.22100067138672
- atom: N7 x: 2.1589999198913574 y:28.683000564575195 z:49.40299987792969
- atom: C5 x: 2.9100000858306885 y:29.089000701904297 z:48.3129997253418
- atom: C6 x: 4.244999885559082 y:28.812999725341797 z:47.972999572753906
- atom: O6 x: 5.076000213623047 y:28.15399932861328 z:48.59700012207031
- atom: N1 x: 4.5980000495910645 y:29.413999557495117 z:46.77299880981445
- atom: C2 x: 3.7769999504089355 y:30.197999954223633 z:46.01499938964844
- atom: N2 x: 4.291999816894531 y:30.69099998474121 z:44.88600158691406
- atom: N3 x: 2.5409998893737793 y:30.482999801635742 z:46.33399963378906
- atom: C4 x: 2.1710000038146973 y:29.89900016784668 z:47.481998443603516
- residue DC has 19 atoms
- atom: P x: -3.7049999237060547 y:29.652999877929688 z:44.64699935913086
- atom: OP1 x: -4.730999946594238 y:30.423999786376953 z:43.893001556396484
- atom: OP2 x: -3.9800000190734863 y:28.243000030517578 z:45.04199981689453
- atom: O5' x: -2.365999937057495 y:29.698999404907227 z:43.7859992980957
- atom: C5' x: -1.8380000591278076 y:30.945999145507812 z:43.33399963378906
- atom: C4' x: -0.75 y:30.716999053955078 z:42.310001373291016
- atom: O4' x: 0.4480000138282776 y:30.246000289916992 z:42.97100067138672
- atom: C3' x: -1.0950000286102295 y:29.672000885009766 z:41.250999450683594
- atom: O3' x: -0.6209999918937683 y:30.075000762939453 z:39.96699905395508
- atom: C2' x: -0.3880000114440918 y:28.417999267578125 z:41.731998443603516
- atom: C1' x: 0.8130000233650208 y:28.95599937438965 z:42.499000549316406
- atom: N1 x: 1.187000036239624 y:28.141000747680664 z:43.66999816894531
- atom: C2 x: 2.503999948501587 y:27.666000366210938 z:43.784000396728516
- atom: O2 x: 3.3299999237060547 y:27.927000045776367 z:42.89099884033203
- atom: N3 x: 2.8429999351501465 y:26.937000274658203 z:44.86800003051758
- atom: C4 x: 1.9320000410079956 y:26.677000045776367 z:45.81399917602539
- atom: N4 x: 2.319999933242798 y:25.972000122070312 z:46.88399887084961
- atom: C5 x: 0.5879999995231628 y:27.132999420166016 z:45.71099853515625
- atom: C6 x: 0.2630000114440918 y:27.854000091552734 z:44.63600158691406
- residue DT has 20 atoms
- atom: P x: -0.828000009059906 y:29.101999282836914 z:38.70800018310547
- atom: OP1 x: -0.8669999837875366 y:29.930999755859375 z:37.46799850463867
- atom: OP2 x: -1.9559999704360962 y:28.173999786376953 z:39.0099983215332
- atom: O5' x: 0.5260000228881836 y:28.270999908447266 z:38.683998107910156
- atom: C5' x: 1.777999997138977 y:28.93600082397461 z:38.53799819946289
- atom: C4' x: 2.8570001125335693 y:27.93600082397461 z:38.20399856567383
- atom: O4' x: 3.250999927520752 y:27.259000778198242 z:39.422000885009766
- atom: C3' x: 2.3949999809265137 y:26.836000442504883 z:37.24700164794922
- atom: O3' x: 3.496999979019165 y:26.400999069213867 z:36.444000244140625
- atom: C2' x: 1.9589999914169312 y:25.736000061035156 z:38.19599914550781
- atom: C1' x: 3.0169999599456787 y:25.868000030517578 z:39.27899932861328
- atom: N1 x: 2.696000099182129 y:25.316999435424805 z:40.617000579833984
- atom: C2 x: 3.7639999389648438 y:24.950000762939453 z:41.40399932861328
- atom: O2 x: 4.928999900817871 y:25.084999084472656 z:41.055999755859375
- atom: N3 x: 3.421999931335449 y:24.41900062561035 z:42.61800003051758
- atom: C4 x: 2.1570000648498535 y:24.222000122070312 z:43.119998931884766
- atom: O4 x: 2.010999917984009 y:23.68400001525879 z:44.21900177001953
- atom: C5 x: 1.0809999704360962 y:24.665000915527344 z:42.263999938964844
- atom: C7 x: -0.328000009059906 y:24.54599952697754 z:42.757999420166016
- atom: C6 x: 1.3980000019073486 y:25.173999786376953 z:41.064998626708984
- residue DG has 22 atoms
- atom: P x: 3.2839999198913574 y:26.125 z:34.874000549316406
- atom: OP1 x: 2.9730000495910645 y:27.427000045776367 z:34.23400115966797
- atom: OP2 x: 2.3570001125335693 y:24.972999572753906 z:34.70800018310547
- atom: O5' x: 4.7270002365112305 y:25.694000244140625 z:34.374000549316406
- atom: C5' x: 5.872000217437744 y:26.429000854492188 z:34.78499984741211
- atom: C4' x: 6.8429999351501465 y:25.516000747680664 z:35.494998931884766
- atom: O4' x: 6.228000164031982 y:24.899999618530273 z:36.65399932861328
- atom: C3' x: 7.355000019073486 y:24.358999252319336 z:34.64799880981445
- atom: O3' x: 8.71399974822998 y:24.148000717163086 z:35.01499938964844
- atom: C2' x: 6.465000152587891 y:23.198999404907227 z:35.07500076293945
- atom: C1' x: 6.324999809265137 y:23.479999542236328 z:36.55500030517578
- atom: N9 x: 5.156000137329102 y:22.92099952697754 z:37.22999954223633
- atom: C8 x: 3.8529999256134033 y:22.968000411987305 z:36.803001403808594
- atom: N7 x: 3.006999969482422 y:22.51300048828125 z:37.68600082397461
- atom: C5 x: 3.7990000247955322 y:22.11199951171875 z:38.75199890136719
- atom: C6 x: 3.437000036239624 y:21.558000564575195 z:40.00899887084961
- atom: O6 x: 2.309999942779541 y:21.3439998626709 z:40.45800018310547
- atom: N1 x: 4.552999973297119 y:21.260000228881836 z:40.77899932861328
- atom: C2 x: 5.854000091552734 y:21.476999282836914 z:40.400001525878906
- atom: N2 x: 6.788000106811523 y:21.107999801635742 z:41.284000396728516
- atom: N3 x: 6.209000110626221 y:22.014999389648438 z:39.24100112915039
- atom: C4 x: 5.132999897003174 y:22.309999465942383 z:38.47100067138672
- residue DA has 21 atoms
- atom: P x: 9.545999526977539 y:22.961999893188477 z:34.34600067138672
- atom: OP1 x: 10.937999725341797 y:23.468000411987305 z:34.21900177001953
- atom: OP2 x: 8.817999839782715 y:22.490999221801758 z:33.14099884033203
- atom: O5' x: 9.5 y:21.81399917602539 z:35.448001861572266
- atom: C5' x: 10.081000328063965 y:22.02400016784668 z:36.736000061035156
- atom: C4' x: 10.329000473022461 y:20.70199966430664 z:37.426998138427734
- atom: O4' x: 9.138999938964844 y:20.261999130249023 z:38.12099838256836
- atom: C3' x: 10.741000175476074 y:19.54400062561035 z:36.52299880981445
- atom: O3' x: 11.741999626159668 y:18.784000396728516 z:37.20500183105469
- atom: C2' x: 9.444000244140625 y:18.773000717163086 z:36.327999114990234
- atom: C1' x: 8.732000350952148 y:18.981000900268555 z:37.65599822998047
- atom: N9 x: 7.264999866485596 y:18.985000610351562 z:37.60900115966797
- atom: C8 x: 6.453999996185303 y:19.448999404907227 z:36.606998443603516
- atom: N7 x: 5.175000190734863 y:19.384000778198242 z:36.88800048828125
- atom: C5 x: 5.138000011444092 y:18.83099937438965 z:38.15700149536133
- atom: C6 x: 4.072999954223633 y:18.517000198364258 z:39.018001556396484
- atom: N6 x: 2.7899999618530273 y:18.732999801635742 z:38.7239990234375
- atom: N1 x: 4.375 y:17.97100067138672 z:40.21099853515625
- atom: C2 x: 5.659999847412109 y:17.757999420166016 z:40.50899887084961
- atom: N3 x: 6.749000072479248 y:18.011999130249023 z:39.78799819946289
- atom: C4 x: 6.415999889373779 y:18.558000564575195 z:38.606998443603516
- residue DT has 20 atoms
- atom: P x: 12.317000389099121 y:17.43600082397461 z:36.55500030517578
- atom: OP1 x: 13.734999656677246 y:17.33300018310547 z:36.98099899291992
- atom: OP2 x: 11.979999542236328 y:17.409000396728516 z:35.11199951171875
- atom: O5' x: 11.479000091552734 y:16.302000045776367 z:37.290000915527344
- atom: C5' x: 11.32800006866455 y:16.334999084472656 z:38.70600128173828
- atom: C4' x: 10.307000160217285 y:15.317999839782715 z:39.159000396728516
- atom: O4' x: 8.970999717712402 y:15.774999618530273 z:38.867000579833984
- atom: C3' x: 10.42199993133545 y:13.913000106811523 z:38.55099868774414
- atom: O3' x: 10.61299991607666 y:12.967000007629395 z:39.61199951171875
- atom: C2' x: 9.071999549865723 y:13.696999549865723 z:37.87799835205078
- atom: C1' x: 8.182999610900879 y:14.621000289916992 z:38.68199920654297
- atom: N1 x: 6.90500020980835 y:15.03499984741211 z:38.07099914550781
- atom: C2 x: 5.755000114440918 y:14.852999687194824 z:38.80699920654297
- atom: O2 x: 5.739999771118164 y:14.33899974822998 z:39.91299819946289
- atom: N3 x: 4.610000133514404 y:15.300000190734863 z:38.196998596191406
- atom: C4 x: 4.497000217437744 y:15.899999618530273 z:36.95600128173828
- atom: O4 x: 3.3940000534057617 y:16.273000717163086 z:36.55699920654297
- atom: C5 x: 5.741000175476074 y:16.042999267578125 z:36.229000091552734
- atom: C7 x: 5.723999977111816 y:16.673999786376953 z:34.87300109863281
- atom: C6 x: 6.864999771118164 y:15.604000091552734 z:36.8129997253418
- residue DG has 22 atoms
- atom: P x: 11.225000381469727 y:11.51099967956543 z:39.29899978637695
- atom: OP1 x: 12.70300006866455 y:11.628000259399414 z:39.255001068115234
- atom: OP2 x: 10.50100040435791 y:10.935999870300293 z:38.132999420166016
- atom: O5' x: 10.819999694824219 y:10.654000282287598 z:40.57899856567383
- atom: C5' x: 10.031000137329102 y:11.215999603271484 z:41.62900161743164
- atom: C4' x: 8.626999855041504 y:10.654999732971191 z:41.597999572753906
- atom: O4' x: 7.829999923706055 y:11.324000358581543 z:40.608001708984375
- atom: C3' x: 8.498000144958496 y:9.168999671936035 z:41.27399826049805
- atom: O3' x: 8.227999687194824 y:8.444999694824219 z:42.47800064086914
- atom: C2' x: 7.328999996185303 y:9.090999603271484 z:40.29499816894531
- atom: C1' x: 6.724999904632568 y:10.484000205993652 z:40.34600067138672
- atom: N9 x: 6.067999839782715 y:10.968999862670898 z:39.12699890136719
- atom: C8 x: 6.690000057220459 y:11.46500015258789 z:38.00699996948242
- atom: N7 x: 5.853000164031982 y:11.904000282287598 z:37.10599899291992
- atom: C5 x: 4.598999977111816 y:11.670999526977539 z:37.6510009765625
- atom: C6 x: 3.306999921798706 y:11.958000183105469 z:37.13800048828125
- atom: O6 x: 2.999000072479248 y:12.503999710083008 z:36.073001861572266
- atom: N1 x: 2.312000036239624 y:11.548999786376953 z:38.00899887084961
- atom: C2 x: 2.5190000534057617 y:10.954000473022461 z:39.215999603271484
- atom: N2 x: 1.4140000343322754 y:10.656999588012695 z:39.900001525878906
- atom: N3 x: 3.7149999141693115 y:10.678999900817871 z:39.7130012512207
- atom: C4 x: 4.704999923706055 y:11.067999839782715 z:38.88600158691406
- residue DC has 19 atoms
- atom: P x: 8.17199993133545 y:6.841000080108643 z:42.446998596191406
- atom: OP1 x: 8.222999572753906 y:6.381999969482422 z:43.845001220703125
- atom: OP2 x: 9.154999732971191 y:6.321000099182129 z:41.47800064086914
- atom: O5' x: 6.704999923706055 y:6.544000148773193 z:41.90299987792969
- atom: C5' x: 5.586999893188477 y:6.679999828338623 z:42.770999908447266
- atom: C4' x: 4.313000202178955 y:6.329999923706055 z:42.04499816894531
- atom: O4' x: 4.117000102996826 y:7.260000228881836 z:40.965999603271484
- atom: C3' x: 4.315999984741211 y:4.946000099182129 z:41.40299987792969
- atom: O3' x: 3.621000051498413 y:4.050000190734863 z:42.27799987792969
- atom: C2' x: 3.617000102996826 y:5.1479997634887695 z:40.055999755859375
- atom: C1' x: 3.2750000953674316 y:6.645999908447266 z:40.01100158691406
- atom: N1 x: 3.4820001125335693 y:7.343999862670898 z:38.71900177001953
- atom: C2 x: 2.378000020980835 y:7.947999954223633 z:38.071998596191406
- atom: O2 x: 1.2549999952316284 y:7.870999813079834 z:38.58700180053711
- atom: N3 x: 2.571000099182129 y:8.600000381469727 z:36.9109992980957
- atom: C4 x: 3.7929999828338623 y:8.67300033569336 z:36.38600158691406
- atom: N4 x: 3.941999912261963 y:9.338000297546387 z:35.24399948120117
- atom: C5 x: 4.923999786376953 y:8.069000244140625 z:37.013999938964844
- atom: C6 x: 4.724999904632568 y:7.420000076293945 z:38.16299819946289
- residue DG has 22 atoms
- atom: P x: 3.5369999408721924 y:2.490000009536743 z:41.92900085449219
- atom: OP1 x: 3.2130000591278076 y:1.7960000038146973 z:43.191001892089844
- atom: OP2 x: 4.730999946594238 y:2.0999999046325684 z:41.15800094604492
- atom: O5' x: 2.25 y:2.365000009536743 z:41.00199890136719
- atom: C5' x: 0.9470000267028809 y:2.680999994277954 z:41.50899887084961
- atom: C4' x: -0.08699999749660492 y:2.5480000972747803 z:40.415000915527344
- atom: O4' x: 0.22200000286102295 y:3.51200008392334 z:39.380001068115234
- atom: C3' x: -0.08799999952316284 y:1.1829999685287476 z:39.724998474121094
- atom: O3' x: -1.4179999828338623 y:0.8140000104904175 z:39.356998443603516
- atom: C2' x: 0.7760000228881836 y:1.4210000038146973 z:38.50400161743164
- atom: C1' x: 0.375 y:2.8369998931884766 z:38.14500045776367
- atom: N9 x: 1.3359999656677246 y:3.5820000171661377 z:37.33599853515625
- atom: C8 x: 2.7079999446868896 y:3.490000009536743 z:37.36399841308594
- atom: N7 x: 3.2909998893737793 y:4.26800012588501 z:36.49399948120117
- atom: C5 x: 2.243000030517578 y:4.920000076293945 z:35.858001708984375
- atom: C6 x: 2.259000062942505 y:5.873000144958496 z:34.814998626708984
- atom: O6 x: 3.2360000610351562 y:6.339000225067139 z:34.21500015258789
- atom: N1 x: 0.9739999771118164 y:6.2789998054504395 z:34.470001220703125
- atom: C2 x: -0.17900000512599945 y:5.818999767303467 z:35.04999923706055
- atom: N2 x: -1.3250000476837158 y:6.327000141143799 z:34.58100128173828
- atom: N3 x: -0.20900000631809235 y:4.921999931335449 z:36.020999908447266
- atom: C4 x: 1.031999945640564 y:4.517000198364258 z:36.37200164794922
- residue DC has 19 atoms
- atom: P x: -1.694000005722046 y:-0.6140000224113464 z:38.65599822998047
- atom: OP1 x: -2.994999885559082 y:-1.1480000019073486 z:39.14899826049805
- atom: OP2 x: -0.46000000834465027 y:-1.430999994277954 z:38.821998596191406
- atom: O5' x: -1.8730000257492065 y:-0.24400000274181366 z:37.11000061035156
- atom: C5' x: -2.808000087738037 y:0.7670000195503235 z:36.698001861572266
- atom: C4' x: -2.8580000400543213 y:0.8840000033378601 z:35.1879997253418
- atom: O4' x: -1.7769999504089355 y:1.7239999771118164 z:34.69900131225586
- atom: C3' x: -2.756999969482422 y:-0.4309999942779541 z:34.39899826049805
- atom: O3' x: -3.569999933242798 y:-0.4269999861717224 z:33.20000076293945
- atom: C2' x: -1.312000036239624 y:-0.4320000112056732 z:33.9370002746582
- atom: C1' x: -1.125 y:1.0440000295639038 z:33.637001037597656
- atom: N1 x: 0.2680000066757202 y:1.524999976158142 z:33.55699920654297
- atom: C2 x: 0.5590000152587891 y:2.6029999256134033 z:32.71099853515625
- atom: O2 x: -0.36800000071525574 y:3.1510000228881836 z:32.09000015258789
- atom: N3 x: 1.8370000123977661 y:3.0220000743865967 z:32.59600067138672
- atom: C4 x: 2.8010001182556152 y:2.4200000762939453 z:33.29399871826172
- atom: N4 x: 4.046999931335449 y:2.8559999465942383 z:33.137001037597656
- atom: C5 x: 2.5299999713897705 y:1.3420000076293945 z:34.178001403808594
- atom: C6 x: 1.2660000324249268 y:0.9300000071525574 z:34.27899932861328
- residue MN has 1 atoms
- atom: MN x: -0.07199999690055847 y:21.77899932861328 z:37.319000244140625
- residue HOH has 1 atoms
- atom: O x: 6.046000003814697 y:3.1649999618530273 z:39.082000732421875
- residue HOH has 1 atoms
- atom: O x: 13.569999694824219 y:10.14799976348877 z:41.25299835205078
- residue HOH has 1 atoms
- atom: O x: -5.609000205993652 y:-0.41600000858306885 z:34.5
- residue HOH has 1 atoms
- atom: O x: 4.633999824523926 y:30.27400016784668 z:40.766998291015625
- residue HOH has 1 atoms
- atom: O x: -5.763999938964844 y:33.71799850463867 z:44.33599853515625
- residue HOH has 1 atoms
- atom: O x: 2.937000036239624 y:31.315000534057617 z:42.645999908447266
- residue HOH has 1 atoms
- atom: O x: 7.23199987411499 y:5.645999908447266 z:38.564998626708984
- chain <Chain id=D> has 9 residues
- residue DG has 23 atoms
- atom: OP3 x: 21.908000946044922 y:-5.788000106811523 z:31.12700080871582
- atom: P x: 22.32200050354004 y:-5.873000144958496 z:29.663000106811523
- atom: OP1 x: 23.104999542236328 y:-4.658999919891357 z:29.17799949645996
- atom: OP2 x: 22.988000869750977 y:-7.199999809265137 z:29.34600067138672
- atom: O5' x: 20.930999755859375 y:-5.881999969482422 z:28.815000534057617
- atom: C5' x: 19.81999969482422 y:-6.675000190734863 z:29.256000518798828
- atom: C4' x: 19.066999435424805 y:-7.238999843597412 z:28.073999404907227
- atom: O4' x: 18.231000900268555 y:-6.247000217437744 z:27.457000732421875
- atom: C3' x: 19.908000946044922 y:-7.807000160217285 z:26.941999435424805
- atom: O3' x: 19.149999618530273 y:-8.842000007629395 z:26.315000534057617
- atom: C2' x: 20.09000015258789 y:-6.60699987411499 z:26.030000686645508
- atom: C1' x: 18.798999786376953 y:-5.822999954223633 z:26.229999542236328
- atom: N9 x: 18.93199920654297 y:-4.373000144958496 z:26.301000595092773
- atom: C8 x: 19.9060001373291 y:-3.635999917984009 z:26.934999465942383
- atom: N7 x: 19.700000762939453 y:-2.3499999046325684 z:26.86199951171875
- atom: C5 x: 18.525999069213867 y:-2.2360000610351562 z:26.134000778198242
- atom: C6 x: 17.79400062561035 y:-1.0950000286102295 z:25.75200080871582
- atom: O6 x: 18.014999389648438 y:0.0860000029206276 z:26.02899932861328
- atom: N1 x: 16.68000030517578 y:-1.4359999895095825 z:24.983999252319336
- atom: C2 x: 16.304000854492188 y:-2.7179999351501465 z:24.660999298095703
- atom: N2 x: 15.206000328063965 y:-2.86299991607666 z:23.91699981689453
- atom: N3 x: 16.96299934387207 y:-3.7890000343322754 z:25.04400062561035
- atom: C4 x: 18.05699920654297 y:-3.4769999980926514 z:25.764999389648438
- residue DT has 20 atoms
- atom: P x: 19.722999572753906 y:-9.595000267028809 z:25.020999908447266
- atom: OP1 x: 19.643999099731445 y:-11.03600025177002 z:25.367000579833984
- atom: OP2 x: 21.0049991607666 y:-9.012999534606934 z:24.559999465942383
- atom: O5' x: 18.625999450683594 y:-9.286999702453613 z:23.91699981689453
- atom: C5' x: 17.94099998474121 y:-8.050000190734863 z:23.905000686645508
- atom: C4' x: 17.011999130249023 y:-7.999000072479248 z:22.722000122070312
- atom: O4' x: 16.64699935913086 y:-6.625 z:22.472999572753906
- atom: C3' x: 17.648000717163086 y:-8.508000373840332 z:21.43199920654297
- atom: O3' x: 16.665000915527344 y:-9.154999732971191 z:20.62299919128418
- atom: C2' x: 18.21299934387207 y:-7.251999855041504 z:20.790000915527344
- atom: C1' x: 17.34600067138672 y:-6.119999885559082 z:21.343000411987305
- atom: N1 x: 18.05500030517578 y:-4.886000156402588 z:21.767000198364258
- atom: C2 x: 17.41699981689453 y:-3.680999994277954 z:21.559999465942383
- atom: O2 x: 16.343000411987305 y:-3.5769999027252197 z:20.979000091552734
- atom: N3 x: 18.089000701904297 y:-2.5910000801086426 z:22.051000595092773
- atom: C4 x: 19.31100082397461 y:-2.578000068664551 z:22.687000274658203
- atom: O4 x: 19.77400016784668 y:-1.5240000486373901 z:23.097999572753906
- atom: C5 x: 19.947999954223633 y:-3.859999895095825 z:22.822999954223633
- atom: C7 x: 21.301000595092773 y:-3.927000045776367 z:23.461000442504883
- atom: C6 x: 19.29599952697754 y:-4.940000057220459 z:22.368000030517578
- residue DC has 19 atoms
- atom: P x: 17.084999084472656 y:-9.819999694824219 z:19.219999313354492
- atom: OP1 x: 16.177000045776367 y:-10.982000350952148 z:18.999000549316406
- atom: OP2 x: 18.55699920654297 y:-10.012999534606934 z:19.167999267578125
- atom: O5' x: 16.697999954223633 y:-8.699999809265137 z:18.16200065612793
- atom: C5' x: 15.357000350952148 y:-8.246999740600586 z:18.04599952697754
- atom: C4' x: 15.291000366210938 y:-7.078000068664551 z:17.093000411987305
- atom: O4' x: 15.998000144958496 y:-5.955999851226807 z:17.679000854492188
- atom: C3' x: 15.972000122070312 y:-7.322999954223633 z:15.744999885559082
- atom: O3' x: 15.348999977111816 y:-6.510000228881836 z:14.746000289916992
- atom: C2' x: 17.37299919128418 y:-6.801000118255615 z:15.98900032043457
- atom: C1' x: 17.03700065612793 y:-5.560999870300293 z:16.798999786376953
- atom: N1 x: 18.117000579833984 y:-4.954999923706055 z:17.600000381469727
- atom: C2 x: 18.066999435424805 y:-3.5810000896453857 z:17.847000122070312
- atom: O2 x: 17.121999740600586 y:-2.9189999103546143 z:17.381999969482422
- atom: N3 x: 19.047000885009766 y:-3.00600004196167 z:18.57200050354004
- atom: C4 x: 20.05699920654297 y:-3.744999885559082 z:19.03499984741211
- atom: N4 x: 21.01300048828125 y:-3.13100004196167 z:19.738000869750977
- atom: C5 x: 20.13599967956543 y:-5.1479997634887695 z:18.79599952697754
- atom: C6 x: 19.152999877929688 y:-5.706999778747559 z:18.086000442504883
- residue DG has 22 atoms
- atom: P x: 15.234000205993652 y:-7.048999786376953 z:13.239999771118164
- atom: OP1 x: 14.206999778747559 y:-8.126999855041504 z:13.246000289916992
- atom: OP2 x: 16.608999252319336 y:-7.3379998207092285 z:12.744999885559082
- atom: O5' x: 14.673999786376953 y:-5.798999786376953 z:12.423999786376953
- atom: C5' x: 13.835000038146973 y:-4.823999881744385 z:13.045000076293945
- atom: C4' x: 14.350000381469727 y:-3.431999921798706 z:12.758999824523926
- atom: O4' x: 15.515000343322754 y:-3.1540000438690186 z:13.57800006866455
- atom: C3' x: 14.777999877929688 y:-3.1989998817443848 z:11.3100004196167
- atom: O3' x: 14.291000366210938 y:-1.9329999685287476 z:10.85200023651123
- atom: C2' x: 16.29599952697754 y:-3.2109999656677246 z:11.37399959564209
- atom: C1' x: 16.589000701904297 y:-2.687999963760376 z:12.770999908447266
- atom: N9 x: 17.82900047302246 y:-3.2009999752044678 z:13.35200023651123
- atom: C8 x: 18.270000457763672 y:-4.50600004196167 z:13.321000099182129
- atom: N7 x: 19.4060001373291 y:-4.679999828338623 z:13.937999725341797
- atom: C5 x: 19.740999221801758 y:-3.4179999828338623 z:14.406999588012695
- atom: C6 x: 20.8700008392334 y:-2.99399995803833 z:15.142999649047852
- atom: O6 x: 21.81599998474121 y:-3.6710000038146973 z:15.54800033569336
- atom: N1 x: 20.833999633789062 y:-1.628999948501587 z:15.395999908447266
- atom: C2 x: 19.836000442504883 y:-0.7789999842643738 z:14.994000434875488
- atom: N2 x: 19.996999740600586 y:0.5090000033378601 z:15.331999778747559
- atom: N3 x: 18.766000747680664 y:-1.1640000343322754 z:14.309000015258789
- atom: C4 x: 18.785999298095703 y:-2.489000082015991 z:14.050000190734863
- residue DG has 22 atoms
- atom: P x: 14.321999549865723 y:-1.5820000171661377 z:9.281999588012695
- atom: OP1 x: 13.01200008392334 y:-0.9649999737739563 z:8.95199966430664
- atom: OP2 x: 14.77400016784668 y:-2.7820000648498535 z:8.529999732971191
- atom: O5' x: 15.46500015258789 y:-0.47600001096725464 z:9.168999671936035
- atom: C5' x: 15.302000045776367 y:0.796999990940094 z:9.78600025177002
- atom: C4' x: 16.645999908447266 y:1.4179999828338623 z:10.090999603271484
- atom: O4' x: 17.479999542236328 y:0.46399998664855957 z:10.789999961853027
- atom: C3' x: 17.46299934387207 y:1.8680000305175781 z:8.876999855041504
- atom: O3' x: 18.23699951171875 y:3.005000114440918 z:9.279000282287598
- atom: C2' x: 18.448999404907227 y:0.7289999723434448 z:8.701000213623047
- atom: C1' x: 18.743999481201172 y:0.4269999861717224 z:10.154000282287598
- atom: N9 x: 19.35700035095215 y:-0.8669999837875366 z:10.437000274658203
- atom: C8 x: 18.961999893188477 y:-2.0980000495910645 z:9.970000267028809
- atom: N7 x: 19.707000732421875 y:-3.0769999027252197 z:10.41100025177002
- atom: C5 x: 20.650999069213867 y:-2.4560000896453857 z:11.215999603271484
- atom: C6 x: 21.711999893188477 y:-3.006999969482422 z:11.972000122070312
- atom: O6 x: 22.041000366210938 y:-4.203999996185303 z:12.086999893188477
- atom: N1 x: 22.422000885009766 y:-2.015000104904175 z:12.651000022888184
- atom: C2 x: 22.142000198364258 y:-0.6679999828338623 z:12.607999801635742
- atom: N2 x: 22.93899917602539 y:0.13600000739097595 z:13.335000038146973
- atom: N3 x: 21.152999877929688 y:-0.1459999978542328 z:11.904999732971191
- atom: C4 x: 20.45199966430664 y:-1.090000033378601 z:11.239999771118164
- residue HOH has 1 atoms
- atom: O x: 18.135000228881836 y:3.9839999675750732 z:12.008999824523926
- residue HOH has 1 atoms
- atom: O x: 13.862000465393066 y:-5.619999885559082 z:23.08300018310547
- residue HOH has 1 atoms
- atom: O x: 21.132999420166016 y:-7.02400016784668 z:13.020999908447266
- residue HOH has 1 atoms
- atom: O x: 24.173999786376953 y:-5.079999923706055 z:33.534000396728516
- chain <Chain id=A> has 427 residues
- residue THR has 7 atoms
- atom: N x: 3.50600004196167 y:-24.027999877929688 z:29.959999084472656
- atom: CA x: 4.690999984741211 y:-23.68400001525879 z:30.798999786376953
- atom: C x: 5.771999835968018 y:-24.729000091552734 z:30.599000930786133
- atom: O x: 6.264999866485596 y:-25.347000122070312 z:31.542999267578125
- atom: CB x: 4.321000099182129 y:-23.61400032043457 z:32.292999267578125
- atom: OG1 x: 4.0879998207092285 y:-24.940000534057617 z:32.81100082397461
- atom: CG2 x: 3.069000005722046 y:-22.75 z:32.46699905395508
- residue LEU has 8 atoms
- atom: N x: 6.114999771118164 y:-24.933000564575195 z:29.336999893188477
- atom: CA x: 7.14900016784668 y:-25.878000259399414 z:28.98900032043457
- atom: C x: 8.472999572753906 y:-25.23200035095215 z:29.381999969482422
- atom: O x: 9.387999534606934 y:-25.909000396728516 z:29.861000061035156
- atom: CB x: 7.109000205993652 y:-26.163999557495117 z:27.488000869750977
- atom: CG x: 5.886000156402588 y:-26.91900062561035 z:26.95199966430664
- atom: CD1 x: 6.002999782562256 y:-27.079999923706055 z:25.44099998474121
- atom: CD2 x: 5.789999961853027 y:-28.275999069213867 z:27.613000869750977
- residue ASN has 8 atoms
- atom: N x: 8.552000045776367 y:-23.913999557495117 z:29.201000213623047
- atom: CA x: 9.74899959564209 y:-23.145999908447266 z:29.527999877929688
- atom: C x: 9.545999526977539 y:-22.253000259399414 z:30.756999969482422
- atom: O x: 10.102999687194824 y:-21.159000396728516 z:30.836000442504883
- atom: CB x: 10.137999534606934 y:-22.27899932861328 z:28.33799934387207
- atom: CG x: 10.458000183105469 y:-23.090999603271484 z:27.111000061035156
- atom: OD1 x: 11.486000061035156 y:-23.759000778198242 z:27.051000595092773
- atom: ND2 x: 9.572999954223633 y:-23.04400062561035 z:26.121000289916992
- residue GLY has 4 atoms
- atom: N x: 8.763999938964844 y:-22.740999221801758 z:31.716999053955078
- atom: CA x: 8.47599983215332 y:-21.97800064086914 z:32.922000885009766
- atom: C x: 9.657999992370605 y:-21.47800064086914 z:33.73500061035156
- atom: O x: 9.645999908447266 y:-20.341999053955078 z:34.20500183105469
- residue GLY has 4 atoms
- atom: N x: 10.67199993133545 y:-22.316999435424805 z:33.91999816894531
- atom: CA x: 11.829999923706055 y:-21.899999618530273 z:34.689998626708984
- atom: C x: 12.550999641418457 y:-20.709999084472656 z:34.073001861572266
- atom: O x: 13.067000389099121 y:-19.834999084472656 z:34.7869987487793
- residue ILE has 8 atoms
- atom: N x: 12.583999633789062 y:-20.67799949645996 z:32.74399948120117
- atom: CA x: 13.246000289916992 y:-19.611000061035156 z:32.01900100708008
- atom: C x: 12.480999946594238 y:-18.308000564575195 z:32.0890007019043
- atom: O x: 13.067999839782715 y:-17.257999420166016 z:32.32899856567383
- atom: CB x: 13.434000015258789 y:-19.965999603271484 z:30.530000686645508
- atom: CG1 x: 14.45300006866455 y:-21.104000091552734 z:30.38800048828125
- atom: CG2 x: 13.859999656677246 y:-18.719999313354492 z:29.7450008392334
- atom: CD1 x: 14.708000183105469 y:-21.54400062561035 z:28.957000732421875
- residue THR has 7 atoms
- atom: N x: 11.17199993133545 y:-18.36400032043457 z:31.886999130249023
- atom: CA x: 10.392000198364258 y:-17.139999389648438 z:31.91200065612793
- atom: C x: 10.234000205993652 y:-16.545000076293945 z:33.3129997253418
- atom: O x: 9.973999977111816 y:-15.357000350952148 z:33.44300079345703
- atom: CB x: 9.015999794006348 y:-17.327999114990234 z:31.190000534057617
- atom: OG1 x: 8.112000465393066 y:-18.10099983215332 z:31.989999771118164
- atom: CG2 x: 9.234999656677246 y:-18.034000396728516 z:29.868000030517578
- residue ASP has 8 atoms
- atom: N x: 10.395000457763672 y:-17.35700035095215 z:34.356998443603516
- atom: CA x: 10.33899974822998 y:-16.850000381469727 z:35.73699951171875
- atom: C x: 11.638999938964844 y:-16.07200050354004 z:35.959999084472656
- atom: O x: 11.659000396728516 y:-14.982999801635742 z:36.53799819946289
- atom: CB x: 10.298999786376953 y:-17.98699951171875 z:36.75400161743164
- atom: CG x: 8.946999549865723 y:-18.631000518798828 z:36.85300064086914
- atom: OD1 x: 7.943999767303467 y:-17.89299964904785 z:36.93299865722656
- atom: OD2 x: 8.88599967956543 y:-19.878000259399414 z:36.87099838256836
- residue MET has 8 atoms
- atom: N x: 12.729999542236328 y:-16.667999267578125 z:35.49599838256836
- atom: CA x: 14.029999732971191 y:-16.049999237060547 z:35.595001220703125
- atom: C x: 13.96500015258789 y:-14.680999755859375 z:34.904998779296875
- atom: O x: 14.3149995803833 y:-13.659000396728516 z:35.48699951171875
- atom: CB x: 15.060999870300293 y:-16.926000595092773 z:34.89899826049805
- atom: CG x: 16.468000411987305 y:-16.416000366210938 z:35.000999450683594
- atom: SD x: 17.45599937438965 y:-17.030000686645508 z:33.643001556396484
- atom: CE x: 18.0049991607666 y:-15.479999542236328 z:32.88600158691406
- residue LEU has 8 atoms
- atom: N x: 13.51099967956543 y:-14.67199993133545 z:33.659000396728516
- atom: CA x: 13.411999702453613 y:-13.447999954223633 z:32.88999938964844
- atom: C x: 12.569999694824219 y:-12.394000053405762 z:33.59400177001953
- atom: O x: 12.833000183105469 y:-11.189000129699707 z:33.472999572753906
- atom: CB x: 12.795000076293945 y:-13.741000175476074 z:31.5310001373291
- atom: CG x: 13.562000274658203 y:-14.668999671936035 z:30.604000091552734
- atom: CD1 x: 12.758000373840332 y:-14.875 z:29.336999893188477
- atom: CD2 x: 14.91100025177002 y:-14.074000358581543 z:30.27899932861328
- residue THR has 7 atoms
- atom: N x: 11.553000450134277 y:-12.848999977111816 z:34.323001861572266
- atom: CA x: 10.657999992370605 y:-11.9399995803833 z:35.02000045776367
- atom: C x: 11.28600025177002 y:-11.27299976348877 z:36.21799850463867
- atom: O x: 11.097000122070312 y:-10.074999809265137 z:36.4109992980957
- atom: CB x: 9.369000434875488 y:-12.640999794006348 z:35.45600128173828
- atom: OG1 x: 8.581999778747559 y:-12.916000366210938 z:34.29399871826172
- atom: CG2 x: 8.564000129699707 y:-11.765999794006348 z:36.387001037597656
- residue GLU has 9 atoms
- atom: N x: 12.020999908447266 y:-12.038000106811523 z:37.02199935913086
- atom: CA x: 12.682999610900879 y:-11.47599983215332 z:38.189998626708984
- atom: C x: 13.690999984741211 y:-10.45199966430664 z:37.67300033569336
- atom: O x: 13.807000160217285 y:-9.345000267028809 z:38.196998596191406
- atom: CB x: 13.37399959564209 y:-12.588000297546387 z:38.98899841308594
- atom: CG x: 14.23799991607666 y:-12.11400032043457 z:40.15599822998047
- atom: CD x: 14.40999984741211 y:-13.187000274658203 z:41.233001708984375
- atom: OE1 x: 14.732999801635742 y:-14.33899974822998 z:40.88600158691406
- atom: OE2 x: 14.22599983215332 y:-12.878000259399414 z:42.43600082397461
- residue LEU has 8 atoms
- atom: N x: 14.407999992370605 y:-10.817999839782715 z:36.62200164794922
- atom: CA x: 15.376999855041504 y:-9.906000137329102 z:36.029998779296875
- atom: C x: 14.710000038146973 y:-8.588000297546387 z:35.63999938964844
- atom: O x: 15.276000022888184 y:-7.519999980926514 z:35.840999603271484
- atom: CB x: 16.006000518798828 y:-10.550999641418457 z:34.801998138427734
- atom: CG x: 17.10099983215332 y:-11.543000221252441 z:35.15599822998047
- atom: CD1 x: 17.541000366210938 y:-12.317999839782715 z:33.928001403808594
- atom: CD2 x: 18.26300048828125 y:-10.758000373840332 z:35.744998931884766
- residue ALA has 5 atoms
- atom: N x: 13.50100040435791 y:-8.6850004196167 z:35.09000015258789
- atom: CA x: 12.727999687194824 y:-7.525000095367432 z:34.66899871826172
- atom: C x: 12.454999923706055 y:-6.618000030517578 z:35.85900115966797
- atom: O x: 12.86400032043457 y:-5.453999996185303 z:35.887001037597656
- atom: CB x: 11.416000366210938 y:-7.9730000495910645 z:34.04499816894531
- residue ASN has 8 atoms
- atom: N x: 11.75100040435791 y:-7.1519999504089355 z:36.84299850463867
- atom: CA x: 11.447999954223633 y:-6.377999782562256 z:38.02799987792969
- atom: C x: 12.694999694824219 y:-5.685999870300293 z:38.54800033569336
- atom: O x: 12.654999732971191 y:-4.507999897003174 z:38.917999267578125
- atom: CB x: 10.857000350952148 y:-7.275000095367432 z:39.09700012207031
- atom: CG x: 9.361000061035156 y:-7.355999946594238 z:38.999000549316406
- atom: OD1 x: 8.668000221252441 y:-6.34499979019165 z:39.154998779296875
- atom: ND2 x: 8.843999862670898 y:-8.548999786376953 z:38.73500061035156
- residue PHE has 11 atoms
- atom: N x: 13.807000160217285 y:-6.414000034332275 z:38.56399917602539
- atom: CA x: 15.053000450134277 y:-5.831999778747559 z:39.0260009765625
- atom: C x: 15.369000434875488 y:-4.565999984741211 z:38.233001708984375
- atom: O x: 15.411999702453613 y:-3.4700000286102295 z:38.79100036621094
- atom: CB x: 16.20800018310547 y:-6.817999839782715 z:38.86600112915039
- atom: CG x: 17.55500030517578 y:-6.203999996185303 z:39.13100051879883
- atom: CD1 x: 18.569000244140625 y:-6.275000095367432 z:38.180999755859375
- atom: CD2 x: 17.791000366210938 y:-5.498000144958496 z:40.30799865722656
- atom: CE1 x: 19.79800033569336 y:-5.644000053405762 z:38.39899826049805
- atom: CE2 x: 19.00200080871582 y:-4.873000144958496 z:40.52799987792969
- atom: CZ x: 20.007999420166016 y:-4.941999912261963 z:39.57600021362305
- residue GLU has 9 atoms
- atom: N x: 15.581000328063965 y:-4.728000164031982 z:36.926998138427734
- atom: CA x: 15.913999557495117 y:-3.611999988555908 z:36.04999923706055
- atom: C x: 14.92199993133545 y:-2.4560000896453857 z:36.1609992980957
- atom: O x: 15.28499984741211 y:-1.3009999990463257 z:35.96900177001953
- atom: CB x: 16.007999420166016 y:-4.086999893188477 z:34.59299850463867
- atom: CG x: 17.152000427246094 y:-5.060999870300293 z:34.27799987792969
- atom: CD x: 18.538000106811523 y:-4.489999771118164 z:34.58700180053711
- atom: OE1 x: 18.69700050354004 y:-3.260999917984009 z:34.5260009765625
- atom: OE2 x: 19.479000091552734 y:-5.265999794006348 z:34.875
- residue LYS has 9 atoms
- atom: N x: 13.677000045776367 y:-2.7669999599456787 z:36.48400115966797
- atom: CA x: 12.642999649047852 y:-1.753000020980835 z:36.61600112915039
- atom: C x: 12.543999671936035 y:-1.121999979019165 z:37.99599838256836
- atom: O x: 12.244000434875488 y:0.06400000303983688 z:38.12799835205078
- atom: CB x: 11.277999877929688 y:-2.3440001010894775 z:36.2859992980957
- atom: CG x: 10.192999839782715 y:-1.8370000123977661 z:37.207000732421875
- atom: CD x: 8.888999938964844 y:-1.6490000486373901 z:36.49800109863281
- atom: CE x: 8.100000381469727 y:-0.546999990940094 z:37.20000076293945
- atom: NZ x: 8.9350004196167 y:0.6930000185966492 z:37.34199905395508
- residue ASN has 8 atoms
- atom: N x: 12.75100040435791 y:-1.9199999570846558 z:39.03200149536133
- atom: CA x: 12.637999534606934 y:-1.3869999647140503 z:40.36899948120117
- atom: C x: 13.968000411987305 y:-0.8429999947547913 z:40.900001525878906
- atom: O x: 14.02299976348877 y:0.24799999594688416 z:41.45899963378906
- atom: CB x: 12.067999839782715 y:-2.4579999446868896 z:41.319000244140625
- atom: CG x: 10.592000007629395 y:-2.7790000438690186 z:41.04800033569336
- atom: OD1 x: 9.795999526977539 y:-1.8949999809265137 z:40.73699951171875
- atom: ND2 x: 10.22700023651123 y:-4.045000076293945 z:41.18899917602539
- residue VAL has 7 atoms
- atom: N x: 15.041999816894531 y:-1.590999960899353 z:40.698001861572266
- atom: CA x: 16.341999053955078 y:-1.1859999895095825 z:41.19900131225586
- atom: C x: 17.093000411987305 y:-0.3190000057220459 z:40.21500015258789
- atom: O x: 17.384000778198242 y:0.828000009059906 z:40.51300048828125
- atom: CB x: 17.204999923706055 y:-2.4189999103546143 z:41.53900146484375
- atom: CG1 x: 18.464000701904297 y:-1.99399995803833 z:42.32699966430664
- atom: CG2 x: 16.385000228881836 y:-3.4179999828338623 z:42.33000183105469
- residue SER has 6 atoms
- atom: N x: 17.406999588012695 y:-0.871999979019165 z:39.04600143432617
- atom: CA x: 18.14699935913086 y:-0.14800000190734863 z:37.999000549316406
- atom: C x: 17.43199920654297 y:1.0440000295639038 z:37.369998931884766
- atom: O x: 18.065000534057617 y:1.8589999675750732 z:36.69300079345703
- atom: CB x: 18.55500030517578 y:-1.0989999771118164 z:36.875
- atom: OG x: 19.535999298095703 y:-2.0239999294281006 z:37.31100082397461
- residue GLN has 9 atoms
- atom: N x: 16.125 y:1.1460000276565552 z:37.58000183105469
- atom: CA x: 15.355999946594238 y:2.244999885559082 z:37.012001037597656
- atom: C x: 15.548999786376953 y:2.385999917984009 z:35.499000549316406
- atom: O x: 15.527999877929688 y:3.4839999675750732 z:34.96200180053711
- atom: CB x: 15.715999603271484 y:3.553999900817871 z:37.7140007019043
- atom: CG x: 15.02299976348877 y:3.7219998836517334 z:39.07899856567383
- atom: CD x: 15.668000221252441 y:4.795000076293945 z:39.96500015258789
- atom: OE1 x: 16.8799991607666 y:4.752999782562256 z:40.2400016784668
- atom: NE2 x: 14.861000061035156 y:5.743000030517578 z:40.42499923706055
- residue ALA has 5 atoms
- atom: N x: 15.746000289916992 y:1.2569999694824219 z:34.82899856567383
- atom: CA x: 15.914999961853027 y:1.1920000314712524 z:33.37699890136719
- atom: C x: 14.652999877929688 y:0.49300000071525574 z:32.85499954223633
- atom: O x: 14.583000183105469 y:-0.7369999885559082 z:32.79999923706055
- atom: CB x: 17.15399932861328 y:0.3790000081062317 z:33.03300094604492
- residue ILE has 8 atoms
- atom: N x: 13.656999588012695 y:1.2949999570846558 z:32.49599838256836
- atom: CA x: 12.378000259399414 y:0.7940000295639038 z:32.0260009765625
- atom: C x: 12.460000038146973 y:0.032999999821186066 z:30.702999114990234
- atom: O x: 11.57699966430664 y:-0.7519999742507935 z:30.37299919128418
- atom: CB x: 11.407999992370605 y:1.9550000429153442 z:31.85700035095215
- atom: CG1 x: 10.034000396728516 y:1.4700000286102295 z:31.36199951171875
- atom: CG2 x: 12.008999824523926 y:2.937000036239624 z:30.867000579833984
- atom: CD1 x: 9.13700008392334 y:0.8830000162124634 z:32.4379997253418
- residue HIS has 10 atoms
- atom: N x: 13.517000198364258 y:0.2529999911785126 z:29.94099998474121
- atom: CA x: 13.626999855041504 y:-0.42500001192092896 z:28.665000915527344
- atom: C x: 14.244000434875488 y:-1.7960000038146973 z:28.760000228881836
- atom: O x: 14.008999824523926 y:-2.6500000953674316 z:27.905000686645508
- atom: CB x: 14.366999626159668 y:0.4620000123977661 z:27.674999237060547
- atom: CG x: 13.645999908447266 y:1.7430000305175781 z:27.393999099731445
- atom: ND1 x: 12.3100004196167 y:1.7760000228881836 z:27.051000595092773
- atom: CD2 x: 14.04800033569336 y:3.0329999923706055 z:27.476999282836914
- atom: CE1 x: 11.918999671936035 y:3.0320000648498535 z:26.941999435424805
- atom: NE2 x: 12.954000473022461 y:3.813999891281128 z:27.19499969482422
- residue LYS has 9 atoms
- atom: N x: 15.031999588012695 y:-2.0220000743865967 z:29.798999786376953
- atom: CA x: 15.621000289916992 y:-3.3359999656677246 z:29.981000900268555
- atom: C x: 14.463000297546387 y:-4.136000156402588 z:30.52199935913086
- atom: O x: 14.262999534606934 y:-5.2829999923706055 z:30.141000747680664
- atom: CB x: 16.773000717163086 y:-3.299999952316284 z:30.996000289916992
- atom: CG x: 17.975000381469727 y:-2.4779999256134033 z:30.540000915527344
- atom: CD x: 19.246000289916992 y:-2.9200000762939453 z:31.253000259399414
- atom: CE x: 20.474000930786133 y:-2.2990000247955322 z:30.60099983215332
- atom: NZ x: 21.74799919128418 y:-2.9100000858306885 z:31.034000396728516
- residue TYR has 12 atoms
- atom: N x: 13.689000129699707 y:-3.5139999389648438 z:31.408000946044922
- atom: CA x: 12.527999877929688 y:-4.175000190734863 z:31.989999771118164
- atom: C x: 11.668999671936035 y:-4.672999858856201 z:30.836999893188477
- atom: O x: 11.380999565124512 y:-5.86899995803833 z:30.731000900268555
- atom: CB x: 11.722999572753906 y:-3.194999933242798 z:32.847999572753906
- atom: CG x: 10.309000015258789 y:-3.6470000743865967 z:33.15999984741211
- atom: CD1 x: 10.064000129699707 y:-4.732999801635742 z:33.992000579833984
- atom: CD2 x: 9.215999603271484 y:-2.9639999866485596 z:32.64899826049805
- atom: CE1 x: 8.758999824523926 y:-5.125 z:34.310001373291016
- atom: CE2 x: 7.9070000648498535 y:-3.3469998836517334 z:32.96200180053711
- atom: CZ x: 7.689000129699707 y:-4.423999786376953 z:33.79100036621094
- atom: OH x: 6.39900016784668 y:-4.795000076293945 z:34.09600067138672
- residue ASN has 8 atoms
- atom: N x: 11.267000198364258 y:-3.75 z:29.965999603271484
- atom: CA x: 10.454999923706055 y:-4.125 z:28.819000244140625
- atom: C x: 11.135000228881836 y:-5.242000102996826 z:28.03499984741211
- atom: O x: 10.5 y:-6.247000217437744 z:27.711999893188477
- atom: CB x: 10.20300006866455 y:-2.9200000762939453 z:27.91200065612793
- atom: CG x: 9.14900016784668 y:-2.003999948501587 z:28.468000411987305
- atom: OD1 x: 8.194000244140625 y:-2.4619998931884766 z:29.083999633789062
- atom: ND2 x: 9.305000305175781 y:-0.7049999833106995 z:28.249000549316406
- residue ALA has 5 atoms
- atom: N x: 12.42199993133545 y:-5.059999942779541 z:27.74799919128418
- atom: CA x: 13.201000213623047 y:-6.046000003814697 z:27.020999908447266
- atom: C x: 13.07800006866455 y:-7.461999893188477 z:27.60099983215332
- atom: O x: 12.82699966430664 y:-8.409000396728516 z:26.867000579833984
- atom: CB x: 14.645999908447266 y:-5.619999885559082 z:26.993999481201172
- residue TYR has 12 atoms
- atom: N x: 13.26200008392334 y:-7.61299991607666 z:28.910999298095703
- atom: CA x: 13.152000427246094 y:-8.927000045776367 z:29.538000106811523
- atom: C x: 11.708000183105469 y:-9.369000434875488 z:29.48699951171875
- atom: O x: 11.395999908447266 y:-10.520000457763672 z:29.2189998626709
- atom: CB x: 13.609999656677246 y:-8.871000289916992 z:31.00200080871582
- atom: CG x: 15.104999542236328 y:-8.961999893188477 z:31.177000045776367
- atom: CD1 x: 15.8100004196167 y:-10.065999984741211 z:30.708999633789062
- atom: CD2 x: 15.821000099182129 y:-7.946000099182129 z:31.804000854492188
- atom: CE1 x: 17.191999435424805 y:-10.159000396728516 z:30.85700035095215
- atom: CE2 x: 17.207000732421875 y:-8.029999732971191 z:31.95599937438965
- atom: CZ x: 17.881999969482422 y:-9.13700008392334 z:31.481000900268555
- atom: OH x: 19.249000549316406 y:-9.23799991607666 z:31.62700080871582
- residue ARG has 11 atoms
- atom: N x: 10.82800006866455 y:-8.425999641418457 z:29.774999618530273
- atom: CA x: 9.402000427246094 y:-8.663000106811523 z:29.78499984741211
- atom: C x: 8.987000465393066 y:-9.234000205993652 z:28.422000885009766
- atom: O x: 8.211000442504883 y:-10.1850004196167 z:28.336999893188477
- atom: CB x: 8.715999603271484 y:-7.334000110626221 z:30.06999969482422
- atom: CG x: 7.258999824523926 y:-7.432000160217285 z:30.378000259399414
- atom: CD x: 7.006999969482422 y:-7.331999778747559 z:31.860000610351562
- atom: NE x: 5.626999855041504 y:-6.922999858856201 z:32.09299850463867
- atom: CZ x: 5.074999809265137 y:-5.839000225067139 z:31.549999237060547
- atom: NH1 x: 5.791999816894531 y:-5.061999797821045 z:30.73699951171875
- atom: NH2 x: 3.812000036239624 y:-5.519999980926514 z:31.82900047302246
- residue LYS has 9 atoms
- atom: N x: 9.53499984741211 y:-8.657999992370605 z:27.354999542236328
- atom: CA x: 9.25 y:-9.10099983215332 z:25.996000289916992
- atom: C x: 9.82699966430664 y:-10.49899959564209 z:25.763999938964844
- atom: O x: 9.098999977111816 y:-11.430000305175781 z:25.424999237060547
- atom: CB x: 9.835000038146973 y:-8.100000381469727 z:24.999000549316406
- atom: CG x: 9.510000228881836 y:-8.36299991607666 z:23.541000366210938
- atom: CD x: 9.970000267028809 y:-7.190000057220459 z:22.68400001525879
- atom: CE x: 10.038000106811523 y:-7.559000015258789 z:21.20800018310547
- atom: NZ x: 10.956999778747559 y:-8.722000122070312 z:20.97100067138672
- residue ALA has 5 atoms
- atom: N x: 11.128999710083008 y:-10.652999877929688 z:25.959999084472656
- atom: CA x: 11.76200008392334 y:-11.949000358581543 z:25.777999877929688
- atom: C x: 10.984999656677246 y:-13.067000389099121 z:26.46299934387207
- atom: O x: 10.824000358581543 y:-14.14799976348877 z:25.902999877929688
- atom: CB x: 13.173999786376953 y:-11.914999961853027 z:26.304000854492188
- residue ALA has 5 atoms
- atom: N x: 10.5 y:-12.812000274658203 z:27.672000885009766
- atom: CA x: 9.753999710083008 y:-13.829999923706055 z:28.402000427246094
- atom: C x: 8.461000442504883 y:-14.230999946594238 z:27.68199920654297
- atom: O x: 8.182000160217285 y:-15.414999961853027 z:27.506999969482422
- atom: CB x: 9.442999839782715 y:-13.34000015258789 z:29.815000534057617
- residue SER has 6 atoms
- atom: N x: 7.676000118255615 y:-13.246000289916992 z:27.266000747680664
- atom: CA x: 6.428999900817871 y:-13.508999824523926 z:26.575000762939453
- atom: C x: 6.638999938964844 y:-14.333000183105469 z:25.29599952697754
- atom: O x: 5.808000087738037 y:-15.168999671936035 z:24.929000854492188
- atom: CB x: 5.763000011444092 y:-12.182000160217285 z:26.229999542236328
- atom: OG x: 4.60099983215332 y:-12.369999885559082 z:25.44099998474121
- residue VAL has 7 atoms
- atom: N x: 7.751999855041504 y:-14.08899974822998 z:24.615999221801758
- atom: CA x: 8.069000244140625 y:-14.795999526977539 z:23.388999938964844
- atom: C x: 8.357000350952148 y:-16.267000198364258 z:23.677000045776367
- atom: O x: 7.839000225067139 y:-17.152999877929688 z:22.985000610351562
- atom: CB x: 9.284000396728516 y:-14.144000053405762 z:22.694000244140625
- atom: CG1 x: 10.064000129699707 y:-15.180999755859375 z:21.92300033569336
- atom: CG2 x: 8.812999725341797 y:-13.031000137329102 z:21.76099967956543
- residue ILE has 8 atoms
- atom: N x: 9.189000129699707 y:-16.523000717163086 z:24.68899917602539
- atom: CA x: 9.541000366210938 y:-17.88800048828125 z:25.08799934387207
- atom: C x: 8.321999549865723 y:-18.58099937438965 z:25.70199966430664
- atom: O x: 8.041000366210938 y:-19.75 z:25.413000106811523
- atom: CB x: 10.6850004196167 y:-17.89900016784668 z:26.131999969482422
- atom: CG1 x: 12.006999969482422 y:-17.5049991607666 z:25.482999801635742
- atom: CG2 x: 10.8100004196167 y:-19.267000198364258 z:26.743000030517578
- atom: CD1 x: 13.130999565124512 y:-17.316999435424805 z:26.476999282836914
- residue ALA has 5 atoms
- atom: N x: 7.605999946594238 y:-17.854999542236328 z:26.55299949645996
- atom: CA x: 6.414000034332275 y:-18.388999938964844 z:27.195999145507812
- atom: C x: 5.311999797821045 y:-18.68000030517578 z:26.170000076293945
- atom: O x: 4.185999870300293 y:-19.033000946044922 z:26.5310001373291
- atom: CB x: 5.906000137329102 y:-17.415000915527344 z:28.23200035095215
- residue LYS has 9 atoms
- atom: N x: 5.626999855041504 y:-18.524999618530273 z:24.888999938964844
- atom: CA x: 4.650000095367432 y:-18.802000045776367 z:23.851999282836914
- atom: C x: 5.250999927520752 y:-19.749000549316406 z:22.81100082397461
- atom: O x: 4.5879998207092285 y:-20.131999969482422 z:21.847999572753906
- atom: CB x: 4.186999797821045 y:-17.496999740600586 z:23.204999923706055
- atom: CG x: 2.924999952316284 y:-17.617000579833984 z:22.378999710083008
- atom: CD x: 2.450000047683716 y:-16.24799919128418 z:21.92099952697754
- atom: CE x: 3.4739999771118164 y:-15.571999549865723 z:21.01799964904785
- atom: NZ x: 3.049999952316284 y:-14.184000015258789 z:20.684999465942383
- residue TYR has 12 atoms
- atom: N x: 6.513000011444092 y:-20.1200008392334 z:23.009000778198242
- atom: CA x: 7.193999767303467 y:-21.051000595092773 z:22.108999252319336
- atom: C x: 6.574999809265137 y:-22.388999938964844 z:22.45599937438965
- atom: O x: 6.429999828338623 y:-22.719999313354492 z:23.631000518798828
- atom: CB x: 8.675000190734863 y:-21.06599998474121 z:22.42099952697754
- atom: CG x: 9.553000450134277 y:-21.915000915527344 z:21.53700065612793
- atom: CD1 x: 9.6850004196167 y:-21.6560001373291 z:20.166000366210938
- atom: CD2 x: 10.39799976348877 y:-22.868000030517578 z:22.111000061035156
- atom: CE1 x: 10.666999816894531 y:-22.320999145507812 z:19.39699935913086
- atom: CE2 x: 11.368000030517578 y:-23.527999877929688 z:21.361000061035156
- atom: CZ x: 11.506999969482422 y:-23.253999710083008 z:20.016000747680664
- atom: OH x: 12.527999877929688 y:-23.892000198364258 z:19.347999572753906
- residue PRO has 7 atoms
- atom: N x: 6.186999797821045 y:-23.167999267578125 z:21.437999725341797
- atom: CA x: 5.563000202178955 y:-24.483999252319336 z:21.61400032043457
- atom: C x: 6.4720001220703125 y:-25.672000885009766 z:21.94700050354004
- atom: O x: 5.98199987411499 y:-26.764999389648438 z:22.20800018310547
- atom: CB x: 4.800000190734863 y:-24.670000076293945 z:20.30299949645996
- atom: CG x: 5.709000110626221 y:-24.02400016784668 z:19.30900001525879
- atom: CD x: 6.133999824523926 y:-22.7450008392334 z:20.025999069213867
- residue HIS has 10 atoms
- atom: N x: 7.7820000648498535 y:-25.472000122070312 z:21.944000244140625
- atom: CA x: 8.687000274658203 y:-26.566999435424805 z:22.27199935913086
- atom: C x: 9.373000144958496 y:-26.291000366210938 z:23.597000122070312
- atom: O x: 9.204000473022461 y:-25.219999313354492 z:24.173999786376953
- atom: CB x: 9.75100040435791 y:-26.72800064086914 z:21.19099998474121
- atom: CG x: 9.192999839782715 y:-26.843000411987305 z:19.81100082397461
- atom: ND1 x: 8.234000205993652 y:-27.770999908447266 z:19.472000122070312
- atom: CD2 x: 9.461999893188477 y:-26.148000717163086 z:18.680999755859375
- atom: CE1 x: 7.934999942779541 y:-27.64299964904785 z:18.191999435424805
- atom: NE2 x: 8.666000366210938 y:-26.665000915527344 z:17.687999725341797
- residue LYS has 9 atoms
- atom: N x: 10.135000228881836 y:-27.26099967956543 z:24.09000015258789
- atom: CA x: 10.866999626159668 y:-27.077999114990234 z:25.336000442504883
- atom: C x: 12.291999816894531 y:-26.701000213623047 z:24.979000091552734
- atom: O x: 13.027999877929688 y:-27.5 z:24.395999908447266
- atom: CB x: 10.859999656677246 y:-28.347000122070312 z:26.18000030517578
- atom: CG x: 12.022000312805176 y:-28.41900062561035 z:27.149999618530273
- atom: CD x: 11.635000228881836 y:-29.1200008392334 z:28.43199920654297
- atom: CE x: 10.968999862670898 y:-28.14699935913086 z:29.37700080871582
- atom: NZ x: 11.902000427246094 y:-27.017000198364258 z:29.71500015258789
- residue ILE has 8 atoms
- atom: N x: 12.661999702453613 y:-25.47100067138672 z:25.326000213623047
- atom: CA x: 13.979999542236328 y:-24.924999237060547 z:25.03700065612793
- atom: C x: 15.112000465393066 y:-25.763999938964844 z:25.604999542236328
- atom: O x: 15.272000312805176 y:-25.878999710083008 z:26.81800079345703
- atom: CB x: 14.07800006866455 y:-23.479999542236328 z:25.554000854492188
- atom: CG1 x: 13.07699966430664 y:-22.618000030517578 z:24.783000946044922
- atom: CG2 x: 15.498000144958496 y:-22.948999404907227 z:25.399999618530273
- atom: CD1 x: 13.050000190734863 y:-21.170000076293945 z:25.19499969482422
- residue LYS has 9 atoms
- atom: N x: 15.897000312805176 y:-26.341999053955078 z:24.70400047302246
- atom: CA x: 17.009000778198242 y:-27.194000244140625 z:25.08300018310547
- atom: C x: 18.363000869750977 y:-26.479000091552734 z:25.097000122070312
- atom: O x: 19.3799991607666 y:-27.075000762939453 z:25.45400047302246
- atom: CB x: 17.05900001525879 y:-28.413999557495117 z:24.1560001373291
- atom: CG x: 15.869000434875488 y:-29.34600067138672 z:24.302000045776367
- atom: CD x: 16.128000259399414 y:-30.667999267578125 z:23.60300064086914
- atom: CE x: 15.015000343322754 y:-31.684999465942383 z:23.892000198364258
- atom: NZ x: 15.324999809265137 y:-33.077999114990234 z:23.42300033569336
- residue SER has 6 atoms
- atom: N x: 18.378000259399414 y:-25.202999114990234 z:24.72100067138672
- atom: CA x: 19.618999481201172 y:-24.429000854492188 z:24.722000122070312
- atom: C x: 19.31399917602539 y:-22.95199966430664 z:24.527000427246094
- atom: O x: 18.201000213623047 y:-22.58300018310547 z:24.15399932861328
- atom: CB x: 20.55699920654297 y:-24.892000198364258 z:23.604000091552734
- atom: OG x: 20.27899932861328 y:-24.198999404907227 z:22.398000717163086
- residue GLY has 4 atoms
- atom: N x: 20.309999465942383 y:-22.110000610351562 z:24.777999877929688
- atom: CA x: 20.1200008392334 y:-20.68199920654297 z:24.617000579833984
- atom: C x: 20.091999053955078 y:-20.316999435424805 z:23.14699935913086
- atom: O x: 19.503999710083008 y:-19.30699920654297 z:22.756000518798828
- residue ALA has 5 atoms
- atom: N x: 20.73200035095215 y:-21.145999908447266 z:22.327999114990234
- atom: CA x: 20.773000717163086 y:-20.899999618530273 z:20.902000427246094
- atom: C x: 19.391000747680664 y:-21.14299964904785 z:20.302000045776367
- atom: O x: 18.961999893188477 y:-20.430999755859375 z:19.392000198364258
- atom: CB x: 21.799999237060547 y:-21.798999786376953 z:20.253000259399414
- residue GLU has 9 atoms
- atom: N x: 18.69300079345703 y:-22.150999069213867 z:20.809999465942383
- atom: CA x: 17.35700035095215 y:-22.451000213623047 z:20.31999969482422
- atom: C x: 16.514999389648438 y:-21.20599937438965 z:20.593000411987305
- atom: O x: 15.798999786376953 y:-20.719999313354492 z:19.715999603271484
- atom: CB x: 16.768999099731445 y:-23.655000686645508 z:21.06100082397461
- atom: CG x: 15.47700023651123 y:-24.17300033569336 z:20.458999633789062
- atom: CD x: 14.970999717712402 y:-25.433000564575195 z:21.138999938964844
- atom: OE1 x: 15.795999526977539 y:-26.31999969482422 z:21.44700050354004
- atom: OE2 x: 13.744999885559082 y:-25.54800033569336 z:21.347999572753906
- residue ALA has 5 atoms
- atom: N x: 16.632999420166016 y:-20.677000045776367 z:21.808000564575195
- atom: CA x: 15.897000312805176 y:-19.48900032043457 z:22.211000442504883
- atom: C x: 16.354000091552734 y:-18.23699951171875 z:21.452999114990234
- atom: O x: 15.574999809265137 y:-17.315000534057617 z:21.225000381469727
- atom: CB x: 16.05699920654297 y:-19.281999588012695 z:23.701000213623047
- residue LYS has 9 atoms
- atom: N x: 17.624000549316406 y:-18.20800018310547 z:21.069000244140625
- atom: CA x: 18.18400001525879 y:-17.07699966430664 z:20.341999053955078
- atom: C x: 17.55699920654297 y:-16.8799991607666 z:18.937999725341797
- atom: O x: 17.700000762939453 y:-15.824000358581543 z:18.320999145507812
- atom: CB x: 19.695999145507812 y:-17.263999938964844 z:20.235000610351562
- atom: CG x: 20.41699981689453 y:-16.229000091552734 z:19.392000198364258
- atom: CD x: 20.399999618530273 y:-14.847999572753906 z:20.023000717163086
- atom: CE x: 21.18199920654297 y:-13.862000465393066 z:19.165000915527344
- atom: NZ x: 22.56599998474121 y:-14.347000122070312 z:18.958999633789062
- residue LYS has 9 atoms
- atom: N x: 16.861000061035156 y:-17.892000198364258 z:18.434999465942383
- atom: CA x: 16.226999282836914 y:-17.773000717163086 z:17.131000518798828
- atom: C x: 15.065999984741211 y:-16.77199935913086 z:17.200000762939453
- atom: O x: 14.831000328063965 y:-16.006999969482422 z:16.256999969482422
- atom: CB x: 15.673999786376953 y:-19.125 z:16.680999755859375
- atom: CG x: 16.683000564575195 y:-20.246999740600586 z:16.594999313354492
- atom: CD x: 15.987000465393066 y:-21.54400062561035 z:16.172000885009766
- atom: CE x: 16.908000946044922 y:-22.753999710083008 z:16.27899932861328
- atom: NZ x: 16.2549991607666 y:-24.025999069213867 z:15.857000350952148
- residue LEU has 8 atoms
- atom: N x: 14.343999862670898 y:-16.790000915527344 z:18.32200050354004
- atom: CA x: 13.184000015258789 y:-15.925999641418457 z:18.53700065612793
- atom: C x: 13.527999877929688 y:-14.432999610900879 z:18.64699935913086
- atom: O x: 14.597000122070312 y:-14.064000129699707 z:19.136999130249023
- atom: CB x: 12.437000274658203 y:-16.3799991607666 z:19.795000076293945
- atom: CG x: 12.102999687194824 y:-17.868000030517578 z:19.90399932861328
- atom: CD1 x: 11.484999656677246 y:-18.17799949645996 z:21.260000228881836
- atom: CD2 x: 11.161999702453613 y:-18.250999450683594 z:18.78499984741211
- residue PRO has 7 atoms
- atom: N x: 12.616000175476074 y:-13.555999755859375 z:18.187000274658203
- atom: CA x: 12.782999992370605 y:-12.100000381469727 z:18.21500015258789
- atom: C x: 12.5 y:-11.564000129699707 z:19.618000030517578
- atom: O x: 11.369999885559082 y:-11.609999656677246 z:20.106000900268555
- atom: CB x: 11.753000259399414 y:-11.631999969482422 z:17.20800018310547
- atom: CG x: 10.605999946594238 y:-12.545000076293945 z:17.520000457763672
- atom: CD x: 11.295999526977539 y:-13.909000396728516 z:17.631999969482422
- residue GLY has 4 atoms
- atom: N x: 13.532999992370605 y:-11.04699993133545 z:20.26300048828125
- atom: CA x: 13.359999656677246 y:-10.550000190734863 z:21.61199951171875
- atom: C x: 14.352999687194824 y:-11.239999771118164 z:22.52199935913086
- atom: O x: 14.593999862670898 y:-10.791000366210938 z:23.634000778198242
- residue VAL has 7 atoms
- atom: N x: 14.920000076293945 y:-12.342000007629395 z:22.047000885009766
- atom: CA x: 15.909000396728516 y:-13.072999954223633 z:22.801000595092773
- atom: C x: 17.238000869750977 y:-12.873000144958496 z:22.11199951171875
- atom: O x: 17.503000259399414 y:-13.482999801635742 z:21.072999954223633
- atom: CB x: 15.623000144958496 y:-14.553999900817871 z:22.81399917602539
- atom: CG1 x: 16.7549991607666 y:-15.277000427246094 z:23.52199935913086
- atom: CG2 x: 14.295000076293945 y:-14.815999984741211 z:23.481000900268555
- residue GLY has 4 atoms
- atom: N x: 18.07200050354004 y:-12.015999794006348 z:22.69700050354004
- atom: CA x: 19.378999710083008 y:-11.729999542236328 z:22.134000778198242
- atom: C x: 20.488000869750977 y:-12.513999938964844 z:22.802000045776367
- atom: O x: 20.222000122070312 y:-13.487000465393066 z:23.506000518798828
- residue THR has 7 atoms
- atom: N x: 21.72800064086914 y:-12.062999725341797 z:22.60300064086914
- atom: CA x: 22.920000076293945 y:-12.718999862670898 z:23.145999908447266
- atom: C x: 22.94099998474121 y:-12.92199993133545 z:24.6560001373291
- atom: O x: 23.170000076293945 y:-14.029999732971191 z:25.128000259399414
- atom: CB x: 24.21299934387207 y:-11.954999923706055 z:22.770000457763672
- atom: OG1 x: 24.249000549316406 y:-11.710000038146973 z:21.356000900268555
- atom: CG2 x: 25.434999465942383 y:-12.774999618530273 z:23.156999588012695
- residue LYS has 9 atoms
- atom: N x: 22.709999084472656 y:-11.857999801635742 z:25.413000106811523
- atom: CA x: 22.726999282836914 y:-11.961000442504883 z:26.865999221801758
- atom: C x: 21.701000213623047 y:-12.954000473022461 z:27.388999938964844
- atom: O x: 22.0 y:-13.739999771118164 z:28.275999069213867
- atom: CB x: 22.475000381469727 y:-10.595999717712402 z:27.5
- atom: CG x: 23.288000106811523 y:-9.480999946594238 z:26.8700008392334
- atom: CD x: 24.777000427246094 y:-9.682999610900879 z:27.093000411987305
- atom: CE x: 25.266000747680664 y:-8.854999542236328 z:28.26300048828125
- atom: NZ x: 25.291000366210938 y:-7.4070000648498535 z:27.913999557495117
- residue ILE has 8 atoms
- atom: N x: 20.489999771118164 y:-12.942000389099121 z:26.85099983215332
- atom: CA x: 19.493000030517578 y:-13.871000289916992 z:27.351999282836914
- atom: C x: 19.766000747680664 y:-15.331000328063965 z:26.969999313354492
- atom: O x: 19.400999069213867 y:-16.253000259399414 z:27.704999923706055
- atom: CB x: 18.073999404907227 y:-13.446999549865723 z:26.920000076293945
- atom: CG1 x: 17.691999435424805 y:-12.152999877929688 z:27.6560001373291
- atom: CG2 x: 17.062999725341797 y:-14.581000328063965 z:27.211999893188477
- atom: CD1 x: 16.2549991607666 y:-11.734999656677246 z:27.492000579833984
- residue ALA has 5 atoms
- atom: N x: 20.427000045776367 y:-15.54800033569336 z:25.836999893188477
- atom: CA x: 20.742000579833984 y:-16.9060001373291 z:25.4060001373291
- atom: C x: 21.82200050354004 y:-17.413000106811523 z:26.351999282836914
- atom: O x: 21.839000701904297 y:-18.582000732421875 z:26.763999938964844
- atom: CB x: 21.256999969482422 y:-16.88800048828125 z:23.981000900268555
- residue GLU has 9 atoms
- atom: N x: 22.719999313354492 y:-16.493000030517578 z:26.68899917602539
- atom: CA x: 23.83099937438965 y:-16.760000228881836 z:27.58300018310547
- atom: C x: 23.28700065612793 y:-17.075000762939453 z:28.95800018310547
- atom: O x: 23.738000869750977 y:-18.0049991607666 z:29.625999450683594
- atom: CB x: 24.726999282836914 y:-15.53600025177002 z:27.64900016784668
- atom: CG x: 25.850000381469727 y:-15.63599967956543 z:28.64299964904785
- atom: CD x: 26.965999603271484 y:-14.670000076293945 z:28.30900001525879
- atom: OE1 x: 27.51799964904785 y:-14.795999526977539 z:27.19700050354004
- atom: OE2 x: 27.283000946044922 y:-13.788999557495117 z:29.141000747680664
- residue LYS has 9 atoms
- atom: N x: 22.308000564575195 y:-16.295000076293945 z:29.381999969482422
- atom: CA x: 21.725000381469727 y:-16.541000366210938 z:30.672000885009766
- atom: C x: 20.95199966430664 y:-17.840999603271484 z:30.590999603271484
- atom: O x: 20.99799919128418 y:-18.648000717163086 z:31.51300048828125
- atom: CB x: 20.82699966430664 y:-15.378999710083008 z:31.089000701904297
- atom: CG x: 21.610000610351562 y:-14.130999565124512 z:31.437000274658203
- atom: CD x: 20.827999114990234 y:-13.210000038146973 z:32.361000061035156
- atom: CE x: 21.672000885009766 y:-12.029999732971191 z:32.82699966430664
- atom: NZ x: 22.187999725341797 y:-11.217000007629395 z:31.687999725341797
- residue ILE has 8 atoms
- atom: N x: 20.256000518798828 y:-18.055999755859375 z:29.479000091552734
- atom: CA x: 19.496000289916992 y:-19.285999298095703 z:29.30500030517578
- atom: C x: 20.44300079345703 y:-20.496999740600586 z:29.367000579833984
- atom: O x: 20.117000579833984 y:-21.52899932861328 z:29.961999893188477
- atom: CB x: 18.714000701904297 y:-19.270000457763672 z:27.961999893188477
- atom: CG1 x: 17.535999298095703 y:-18.297000885009766 z:28.06399917602539
- atom: CG2 x: 18.194000244140625 y:-20.665000915527344 z:27.6299991607666
- atom: CD1 x: 16.844999313354492 y:-18.011999130249023 z:26.757999420166016
- residue ASP has 8 atoms
- atom: N x: 21.618000030517578 y:-20.37299919128418 z:28.761999130249023
- atom: CA x: 22.56999969482422 y:-21.466999053955078 z:28.795000076293945
- atom: C x: 22.993999481201172 y:-21.802000045776367 z:30.23200035095215
- atom: O x: 23.159000396728516 y:-22.97800064086914 z:30.573999404907227
- atom: CB x: 23.785999298095703 y:-21.145999908447266 z:27.93199920654297
- atom: CG x: 23.558000564575195 y:-21.47599983215332 z:26.459999084472656
- atom: OD1 x: 22.81999969482422 y:-22.45199966430664 z:26.187999725341797
- atom: OD2 x: 24.12299919128418 y:-20.783000946044922 z:25.57900047302246
- residue GLU has 9 atoms
- atom: N x: 23.165000915527344 y:-20.785999298095703 z:31.07900047302246
- atom: CA x: 23.533000946044922 y:-21.05699920654297 z:32.4640007019043
- atom: C x: 22.351999282836914 y:-21.722000122070312 z:33.15700149536133
- atom: O x: 22.476999282836914 y:-22.834999084472656 z:33.65800094604492
- atom: CB x: 23.89900016784668 y:-19.783000946044922 z:33.233001708984375
- atom: CG x: 24.15999984741211 y:-20.05900001525879 z:34.72200012207031
- atom: CD x: 24.485000610351562 y:-18.812999725341797 z:35.542999267578125
- atom: OE1 x: 25.302000045776367 y:-17.989999771118164 z:35.09000015258789
- atom: OE2 x: 23.93899917602539 y:-18.665000915527344 z:36.654998779296875
- residue PHE has 11 atoms
- atom: N x: 21.201000213623047 y:-21.04800033569336 z:33.17599868774414
- atom: CA x: 20.01799964904785 y:-21.60300064086914 z:33.83700180053711
- atom: C x: 19.79599952697754 y:-23.05900001525879 z:33.45500183105469
- atom: O x: 19.496999740600586 y:-23.89900016784668 z:34.303001403808594
- atom: CB x: 18.76099967956543 y:-20.81599998474121 z:33.494998931884766
- atom: CG x: 17.554000854492188 y:-21.259000778198242 z:34.27399826049805
- atom: CD1 x: 17.402999877929688 y:-20.89900016784668 z:35.61000061035156
- atom: CD2 x: 16.577999114990234 y:-22.05699920654297 z:33.68299865722656
- atom: CE1 x: 16.29599952697754 y:-21.326000213623047 z:36.34700012207031
- atom: CE2 x: 15.472000122070312 y:-22.486000061035156 z:34.4119987487793
- atom: CZ x: 15.331999778747559 y:-22.1200008392334 z:35.74599838256836
- residue LEU has 8 atoms
- atom: N x: 19.929000854492188 y:-23.354999542236328 z:32.16999816894531
- atom: CA x: 19.76099967956543 y:-24.719999313354492 z:31.724000930786133
- atom: C x: 20.87299919128418 y:-25.54400062561035 z:32.38100051879883
- atom: O x: 20.597000122070312 y:-26.49799919128418 z:33.11199951171875
- atom: CB x: 19.84000015258789 y:-24.78700065612793 z:30.19499969482422
- atom: CG x: 18.53700065612793 y:-25.14900016784668 z:29.46299934387207
- atom: CD1 x: 17.368999481201172 y:-24.40399932861328 z:30.06100082397461
- atom: CD2 x: 18.67099952697754 y:-24.833999633789062 z:27.979999542236328
- residue ALA has 5 atoms
- atom: N x: 22.125 y:-25.1560001373291 z:32.15299987792969
- atom: CA x: 23.2549991607666 y:-25.881000518798828 z:32.73099899291992
- atom: C x: 23.19700050354004 y:-25.913000106811523 z:34.263999938964844
- atom: O x: 22.674999237060547 y:-26.856000900268555 z:34.8650016784668
- atom: CB x: 24.572999954223633 y:-25.256999969482422 z:32.26499938964844
- residue THR has 7 atoms
- atom: N x: 23.732999801635742 y:-24.874000549316406 z:34.891998291015625
- atom: CA x: 23.746999740600586 y:-24.791000366210938 z:36.345001220703125
- atom: C x: 22.393999099731445 y:-25.16200065612793 z:36.95800018310547
- atom: O x: 22.26799964904785 y:-26.194000244140625 z:37.60900115966797
- atom: CB x: 24.17799949645996 y:-23.371000289916992 z:36.810001373291016
- atom: OG1 x: 23.086000442504883 y:-22.448999404907227 z:36.68899917602539
- atom: CG2 x: 25.349000930786133 y:-22.87700080871582 z:35.946998596191406
- residue GLY has 4 atoms
- atom: N x: 21.3799991607666 y:-24.339000701904297 z:36.72800064086914
- atom: CA x: 20.069000244140625 y:-24.611000061035156 z:37.29199981689453
- atom: C x: 19.61400032043457 y:-23.41699981689453 z:38.10900115966797
- atom: O x: 18.61400032043457 y:-23.454999923706055 z:38.83599853515625
- residue LYS has 9 atoms
- atom: N x: 20.375999450683594 y:-22.34000015258789 z:37.97700119018555
- atom: CA x: 20.104999542236328 y:-21.104000091552734 z:38.680999755859375
- atom: C x: 20.933000564575195 y:-20.047000885009766 z:37.987998962402344
- atom: O x: 21.757999420166016 y:-20.365999221801758 z:37.137001037597656
- atom: CB x: 20.530000686645508 y:-21.229999542236328 z:40.150001525878906
- atom: CG x: 22.016000747680664 y:-21.55299949645996 z:40.37799835205078
- atom: CD x: 22.30500030517578 y:-21.792999267578125 z:41.87099838256836
- atom: CE x: 23.71500015258789 y:-22.350000381469727 z:42.15399932861328
- atom: NZ x: 24.823999404907227 y:-21.365999221801758 z:41.97200012207031
- residue LEU has 8 atoms
- atom: N x: 20.711000442504883 y:-18.78700065612793 z:38.33000183105469
- atom: CA x: 21.5 y:-17.73200035095215 z:37.724998474121094
- atom: C x: 22.31999969482422 y:-17.052000045776367 z:38.80699920654297
- atom: O x: 21.7810001373291 y:-16.632999420166016 z:39.83000183105469
- atom: CB x: 20.608999252319336 y:-16.709999084472656 z:37.027000427246094
- atom: CG x: 21.32699966430664 y:-15.911999702453613 z:35.935001373291016
- atom: CD1 x: 21.85099983215332 y:-16.854000091552734 z:34.85599899291992
- atom: CD2 x: 20.36400032043457 y:-14.904999732971191 z:35.32699966430664
- residue ARG has 11 atoms
- atom: N x: 23.6299991607666 y:-16.968000411987305 z:38.57899856567383
- atom: CA x: 24.55500030517578 y:-16.33300018310547 z:39.516998291015625
- atom: C x: 24.148000717163086 y:-14.880000114440918 z:39.749000549316406
- atom: O x: 24.016000747680664 y:-14.442000389099121 z:40.891998291015625
- atom: CB x: 25.979000091552734 y:-16.405000686645508 z:38.9640007019043
- atom: CG x: 26.475000381469727 y:-17.833999633789062 z:38.73099899291992
- atom: CD x: 27.805999755859375 y:-17.840999603271484 z:37.98899841308594
- atom: NE x: 27.750999450683594 y:-16.979000091552734 z:36.808998107910156
- atom: CZ x: 28.722999572753906 y:-16.86400032043457 z:35.904998779296875
- atom: NH1 x: 29.840999603271484 y:-17.56399917602539 z:36.03200149536133
- atom: NH2 x: 28.58300018310547 y:-16.033000946044922 z:34.875999450683594
- residue LYS has 9 atoms
- atom: N x: 23.94099998474121 y:-14.14799976348877 z:38.65700149536133
- atom: CA x: 23.523000717163086 y:-12.746999740600586 z:38.71799850463867
- atom: C x: 22.263999938964844 y:-12.520000457763672 z:39.571998596191406
- atom: O x: 22.094999313354492 y:-11.446999549865723 z:40.1609992980957
- atom: CB x: 23.275999069213867 y:-12.211000442504883 z:37.30699920654297
- atom: CG x: 22.777999877929688 y:-10.774999618530273 z:37.26499938964844
- atom: CD x: 22.426000595092773 y:-10.364999771118164 z:35.8489990234375
- atom: CE x: 21.812000274658203 y:-8.97700023651123 z:35.79999923706055
- atom: NZ x: 22.783000946044922 y:-7.9079999923706055 z:36.16999816894531
- residue LEU has 8 atoms
- atom: N x: 21.37700080871582 y:-13.51099967956543 z:39.630001068115234
- atom: CA x: 20.15399932861328 y:-13.38599967956543 z:40.43600082397461
- atom: C x: 20.409000396728516 y:-13.715999603271484 z:41.909000396728516
- atom: O x: 19.76099967956543 y:-13.151000022888184 z:42.79199981689453
- atom: CB x: 19.0310001373291 y:-14.29800033569336 z:39.91299819946289
- atom: CG x: 18.204999923706055 y:-13.871000289916992 z:38.69599914550781
- atom: CD1 x: 17.20599937438965 y:-14.956000328063965 z:38.374000549316406
- atom: CD2 x: 17.489999771118164 y:-12.565999984741211 z:38.97100067138672
- residue GLU has 9 atoms
- atom: N x: 21.33099937438965 y:-14.642000198364258 z:42.17399978637695
- atom: CA x: 21.6560001373291 y:-14.987000465393066 z:43.553001403808594
- atom: C x: 22.179000854492188 y:-13.70199966430664 z:44.17900085449219
- atom: O x: 21.764999389648438 y:-13.3149995803833 z:45.27199935913086
- atom: CB x: 22.73900032043457 y:-16.07699966430664 z:43.624000549316406
- atom: CG x: 22.267000198364258 y:-17.479999542236328 z:43.24599838256836
- atom: CD x: 21.235000610351562 y:-18.04599952697754 z:44.209999084472656
- atom: OE1 x: 20.523000717163086 y:-18.98900032043457 z:43.81399917602539
- atom: OE2 x: 21.13599967956543 y:-17.562999725341797 z:45.36000061035156
- residue LYS has 9 atoms
- atom: N x: 23.073999404907227 y:-13.038000106811523 z:43.45399856567383
- atom: CA x: 23.663999557495117 y:-11.78600025177002 z:43.90700149536133
- atom: C x: 22.57200050354004 y:-10.781999588012695 z:44.24700164794922
- atom: O x: 22.555999755859375 y:-10.211999893188477 z:45.334999084472656
- atom: CB x: 24.594999313354492 y:-11.241999626159668 z:42.821998596191406
- atom: CG x: 25.749000549316406 y:-12.201000213623047 z:42.54100036621094
- atom: CD x: 26.69700050354004 y:-11.706000328063965 z:41.46500015258789
- atom: CE x: 27.738000869750977 y:-12.774999618530273 z:41.16299819946289
- atom: NZ x: 28.597999572753906 y:-12.456000328063965 z:39.9900016784668
- residue ILE has 8 atoms
- atom: N x: 21.650999069213867 y:-10.579999923706055 z:43.3129997253418
- atom: CA x: 20.538999557495117 y:-9.666999816894531 z:43.53300094604492
- atom: C x: 19.79400062561035 y:-10.114999771118164 z:44.78499984741211
- atom: O x: 19.698999404907227 y:-9.368000030517578 z:45.75299835205078
- atom: CB x: 19.576000213623047 y:-9.663000106811523 z:42.31800079345703
- atom: CG1 x: 20.308000564575195 y:-9.105999946594238 z:41.09299850463867
- atom: CG2 x: 18.336999893188477 y:-8.845999717712402 z:42.625
- atom: CD1 x: 19.481000900268555 y:-9.064000129699707 z:39.84400177001953
- residue ARG has 11 atoms
- atom: N x: 19.283000946044922 y:-11.342000007629395 z:44.7599983215332
- atom: CA x: 18.551000595092773 y:-11.920999526977539 z:45.88999938964844
- atom: C x: 19.215999603271484 y:-11.687000274658203 z:47.249000549316406
- atom: O x: 18.5310001373291 y:-11.527000427246094 z:48.262001037597656
- atom: CB x: 18.378999710083008 y:-13.430000305175781 z:45.68199920654297
- atom: CG x: 17.430999755859375 y:-13.803000450134277 z:44.56800079345703
- atom: CD x: 17.413000106811523 y:-15.305000305175781 z:44.323001861572266
- atom: NE x: 16.273000717163086 y:-15.689000129699707 z:43.492000579833984
- atom: CZ x: 15.977999687194824 y:-16.937000274658203 z:43.132999420166016
- atom: NH1 x: 16.740999221801758 y:-17.950000762939453 z:43.525001525878906
- atom: NH2 x: 14.906000137329102 y:-17.176000595092773 z:42.38800048828125
- residue GLN has 9 atoms
- atom: N x: 20.548999786376953 y:-11.678999900817871 z:47.263999938964844
- atom: CA x: 21.320999145507812 y:-11.486000061035156 z:48.492000579833984
- atom: C x: 21.540000915527344 y:-10.038999557495117 z:48.88999938964844
- atom: O x: 21.20199966430664 y:-9.645999908447266 z:50.005001068115234
- atom: CB x: 22.667999267578125 y:-12.187999725341797 z:48.374000549316406
- atom: CG x: 22.65399932861328 y:-13.574000358581543 z:48.97700119018555
- atom: CD x: 23.506999969482422 y:-14.543000221252441 z:48.20399856567383
- atom: OE1 x: 24.591999053955078 y:-14.189000129699707 z:47.731998443603516
- atom: NE2 x: 23.027000427246094 y:-15.782999992370605 z:48.07099914550781
- residue ASP has 8 atoms
- atom: N x: 22.11400032043457 y:-9.253000259399414 z:47.983001708984375
- atom: CA x: 22.3700008392334 y:-7.834000110626221 z:48.22999954223633
- atom: C x: 21.216999053955078 y:-7.169000148773193 z:48.99300003051758
- atom: O x: 20.115999221801758 y:-7.050000190734863 z:48.47700119018555
- atom: CB x: 22.594999313354492 y:-7.125999927520752 z:46.89099884033203
- atom: CG x: 22.950000762939453 y:-5.663000106811523 z:47.05400085449219
- atom: OD1 x: 23.440000534057617 y:-5.063000202178955 z:46.07400131225586
- atom: OD2 x: 22.735000610351562 y:-5.116000175476074 z:48.15800094604492
- residue ASP has 8 atoms
- atom: N x: 21.47599983215332 y:-6.7230000495910645 z:50.21699905395508
- atom: CA x: 20.43600082397461 y:-6.09499979019165 z:51.03499984741211
- atom: C x: 19.680999755859375 y:-4.929999828338623 z:50.38100051879883
- atom: O x: 18.44499969482422 y:-4.921999931335449 z:50.34199905395508
- atom: CB x: 21.023000717163086 y:-5.610000133514404 z:52.36600112915039
- atom: CG x: 20.0 y:-4.864999771118164 z:53.21699905395508
- atom: OD1 x: 19.062999725341797 y:-5.508999824523926 z:53.75
- atom: OD2 x: 20.125 y:-3.630000114440918 z:53.34400177001953
- residue THR has 7 atoms
- atom: N x: 20.41900062561035 y:-3.941999912261963 z:49.88199996948242
- atom: CA x: 19.788000106811523 y:-2.7799999713897705 z:49.266998291015625
- atom: C x: 18.878999710083008 y:-3.131999969482422 z:48.10300064086914
- atom: O x: 17.701000213623047 y:-2.7730000019073486 z:48.104000091552734
- atom: CB x: 20.826000213623047 y:-1.7740000486373901 z:48.766998291015625
- atom: OG1 x: 21.684999465942383 y:-1.3969999551773071 z:49.84700012207031
- atom: CG2 x: 20.131000518798828 y:-0.531000018119812 z:48.229000091552734
- residue SER has 6 atoms
- atom: N x: 19.43400001525879 y:-3.8239998817443848 z:47.11199951171875
- atom: CA x: 18.676000595092773 y:-4.2210001945495605 z:45.930999755859375
- atom: C x: 17.445999145507812 y:-4.991000175476074 z:46.375999450683594
- atom: O x: 16.354000091552734 y:-4.8429999351501465 z:45.827999114990234
- atom: CB x: 19.527999877929688 y:-5.125999927520752 z:45.04399871826172
- atom: OG x: 20.857999801635742 y:-4.6479997634887695 z:44.95500183105469
- residue SER has 6 atoms
- atom: N x: 17.650999069213867 y:-5.809000015258789 z:47.39699935913086
- atom: CA x: 16.614999771118164 y:-6.6579999923706055 z:47.946998596191406
- atom: C x: 15.543000221252441 y:-5.864999771118164 z:48.672000885009766
- atom: O x: 14.366999626159668 y:-6.239999771118164 z:48.676998138427734
- atom: CB x: 17.256999969482422 y:-7.659999847412109 z:48.895999908447266
- atom: OG x: 16.29199981689453 y:-8.527000427246094 z:49.439998626708984
- residue SER has 6 atoms
- atom: N x: 15.956999778747559 y:-4.765999794006348 z:49.28799819946289
- atom: CA x: 15.032999992370605 y:-3.927000045776367 z:50.02299880981445
- atom: C x: 14.21399974822998 y:-3.0350000858306885 z:49.106998443603516
- atom: O x: 13.001999855041504 y:-2.947000026702881 z:49.257999420166016
- atom: CB x: 15.791999816894531 y:-3.0929999351501465 z:51.06100082397461
- atom: OG x: 16.2549991607666 y:-3.9210000038146973 z:52.12300109863281
- residue ILE has 8 atoms
- atom: N x: 14.859999656677246 y:-2.371999979019165 z:48.159000396728516
- atom: CA x: 14.121000289916992 y:-1.5190000534057617 z:47.24399948120117
- atom: C x: 13.07699966430664 y:-2.390000104904175 z:46.57600021362305
- atom: O x: 11.907999992370605 y:-2.01200008392334 z:46.41999816894531
- atom: CB x: 15.031999588012695 y:-0.9359999895095825 z:46.16899871826172
- atom: CG1 x: 16.11400032043457 y:-0.07800000160932541 z:46.83300018310547
- atom: CG2 x: 14.199000358581543 y:-0.11400000005960464 z:45.18199920654297
- atom: CD1 x: 17.23699951171875 y:0.3479999899864197 z:45.91999816894531
- residue ASN has 8 atoms
- atom: N x: 13.520000457763672 y:-3.5810000896453857 z:46.20199966430664
- atom: CA x: 12.65999984741211 y:-4.548999786376953 z:45.5620002746582
- atom: C x: 11.432000160217285 y:-4.789000034332275 z:46.38999938964844
- atom: O x: 10.333999633789062 y:-4.831999778747559 z:45.874000549316406
- atom: CB x: 13.383999824523926 y:-5.864999771118164 z:45.374000549316406
- atom: CG x: 13.378999710083008 y:-6.310999870300293 z:43.948001861572266
- atom: OD1 x: 12.390000343322754 y:-6.114999771118164 z:43.23500061035156
- atom: ND2 x: 14.47599983215332 y:-6.923999786376953 z:43.51100158691406
- residue PHE has 11 atoms
- atom: N x: 11.60099983215332 y:-4.936999797821045 z:47.6879997253418
- atom: CA x: 10.439000129699707 y:-5.195000171661377 z:48.50699996948242
- atom: C x: 9.46399974822998 y:-4.026000022888184 z:48.55799865722656
- atom: O x: 8.288000106811523 y:-4.184000015258789 z:48.26900100708008
- atom: CB x: 10.843999862670898 y:-5.577000141143799 z:49.92599868774414
- atom: CG x: 9.680000305175781 y:-5.688000202178955 z:50.8489990234375
- atom: CD1 x: 8.789999961853027 y:-6.736999988555908 z:50.729000091552734
- atom: CD2 x: 9.430999755859375 y:-4.703000068664551 z:51.792999267578125
- atom: CE1 x: 7.666999816894531 y:-6.803999900817871 z:51.534000396728516
- atom: CE2 x: 8.303000450134277 y:-4.763999938964844 z:52.60100173950195
- atom: CZ x: 7.425000190734863 y:-5.815000057220459 z:52.47100067138672
- residue LEU has 8 atoms
- atom: N x: 9.954999923706055 y:-2.8559999465942383 z:48.935001373291016
- atom: CA x: 9.11299991607666 y:-1.6629999876022339 z:49.029998779296875
- atom: C x: 8.187000274658203 y:-1.444000005722046 z:47.8489990234375
- atom: O x: 6.998000144958496 y:-1.2089999914169312 z:48.0359992980957
- atom: CB x: 9.97599983215332 y:-0.42399999499320984 z:49.19599914550781
- atom: CG x: 10.741999626159668 y:-0.367000013589859 z:50.50699996948242
- atom: CD1 x: 11.949000358581543 y:0.5139999985694885 z:50.32600021362305
- atom: CD2 x: 9.82699966430664 y:0.1289999932050705 z:51.612998962402344
- residue THR has 7 atoms
- atom: N x: 8.727999687194824 y:-1.5180000066757202 z:46.63600158691406
- atom: CA x: 7.913000106811523 y:-1.3020000457763672 z:45.45000076293945
- atom: C x: 6.660999774932861 y:-2.1670000553131104 z:45.41999816894531
- atom: O x: 5.74399995803833 y:-1.86899995803833 z:44.67499923706055
- atom: CB x: 8.699999809265137 y:-1.5509999990463257 z:44.16999816894531
- atom: OG1 x: 8.991000175476074 y:-2.943000078201294 z:44.060001373291016
- atom: CG2 x: 9.998000144958496 y:-0.7699999809265137 z:44.17599868774414
- residue ARG has 11 atoms
- atom: N x: 6.604000091552734 y:-3.2290000915527344 z:46.2239990234375
- atom: CA x: 5.4070000648498535 y:-4.083000183105469 z:46.25
- atom: C x: 4.270999908447266 y:-3.384000062942505 z:46.97600173950195
- atom: O x: 3.1610000133514404 y:-3.9130001068115234 z:47.06999969482422
- atom: CB x: 5.684999942779541 y:-5.426000118255615 z:46.926998138427734
- atom: CG x: 6.485000133514404 y:-6.410999774932861 z:46.06700134277344
- atom: CD x: 7.070000171661377 y:-7.552000045776367 z:46.89799880981445
- atom: NE x: 6.085000038146973 y:-8.112000465393066 z:47.81800079345703
- atom: CZ x: 6.336999893188477 y:-9.085000038146973 z:48.68600082397461
- atom: NH1 x: 7.546000003814697 y:-9.621999740600586 z:48.762001037597656
- atom: NH2 x: 5.382999897003174 y:-9.510000228881836 z:49.492000579833984
- residue VAL has 7 atoms
- atom: N x: 4.554999828338623 y:-2.2019999027252197 z:47.51300048828125
- atom: CA x: 3.5380001068115234 y:-1.4140000343322754 z:48.19200134277344
- atom: C x: 3.002000093460083 y:-0.4729999899864197 z:47.124000549316406
- atom: O x: 3.7679998874664307 y:0.1459999978542328 z:46.38100051879883
- atom: CB x: 4.129000186920166 y:-0.6010000109672546 z:49.367000579833984
- atom: CG1 x: 3.115000009536743 y:0.40299999713897705 z:49.87699890136719
- atom: CG2 x: 4.514999866485596 y:-1.5449999570846558 z:50.49800109863281
- residue SER has 6 atoms
- atom: N x: 1.6820000410079956 y:-0.3880000114440918 z:47.029998779296875
- atom: CA x: 1.0529999732971191 y:0.453000009059906 z:46.0260009765625
- atom: C x: 1.2309999465942383 y:1.902999997138977 z:46.388999938964844
- atom: O x: 0.7540000081062317 y:2.3459999561309814 z:47.4370002746582
- atom: CB x: -0.4350000023841858 y:0.12800000607967377 z:45.922000885009766
- atom: OG x: -0.9639999866485596 y:0.6510000228881836 z:44.733001708984375
- residue GLY has 4 atoms
- atom: N x: 1.9040000438690186 y:2.6449999809265137 z:45.51599884033203
- atom: CA x: 2.1429998874664307 y:4.053999900817871 z:45.7760009765625
- atom: C x: 3.634999990463257 y:4.316999912261963 z:45.90299987792969
- atom: O x: 4.10099983215332 y:5.461999893188477 z:45.915000915527344
- residue ILE has 8 atoms
- atom: N x: 4.392000198364258 y:3.2300000190734863 z:45.9900016784668
- atom: CA x: 5.824999809265137 y:3.325000047683716 z:46.11600112915039
- atom: C x: 6.491000175476074 y:2.7769999504089355 z:44.880001068115234
- atom: O x: 6.464000225067139 y:1.5800000429153442 z:44.64099884033203
- atom: CB x: 6.308000087738037 y:2.569000005722046 z:47.356998443603516
- atom: CG1 x: 5.689000129699707 y:3.2200000286102295 z:48.59600067138672
- atom: CG2 x: 7.823999881744385 y:2.5880000591278076 z:47.42300033569336
- atom: CD1 x: 5.960000038146973 y:2.489000082015991 z:49.89400100708008
- residue GLY has 4 atoms
- atom: N x: 7.071000099182129 y:3.6670000553131104 z:44.08700180053711
- atom: CA x: 7.74399995803833 y:3.2360000610351562 z:42.8849983215332
- atom: C x: 9.239999771118164 y:3.2160000801086426 z:43.108001708984375
- atom: O x: 9.706999778747559 y:3.369999885559082 z:44.23500061035156
- residue PRO has 7 atoms
- atom: N x: 10.027999877929688 y:3.0280001163482666 z:42.04600143432617
- atom: CA x: 11.480999946594238 y:3.003999948501587 z:42.20600128173828
- atom: C x: 11.973999977111816 y:4.1529998779296875 z:43.07600021362305
- atom: O x: 12.366000175476074 y:3.944999933242798 z:44.2140007019043
- atom: CB x: 11.97599983215332 y:3.0999999046325684 z:40.766998291015625
- atom: CG x: 10.942999839782715 y:2.306999921798706 z:40.03300094604492
- atom: CD x: 9.645000457763672 y:2.819000005722046 z:40.638999938964844
- residue SER has 6 atoms
- atom: N x: 11.925999641418457 y:5.367000102996826 z:42.54199981689453
- atom: CA x: 12.390000343322754 y:6.557000160217285 z:43.24700164794922
- atom: C x: 12.029000282287598 y:6.611000061035156 z:44.720001220703125
- atom: O x: 12.913999557495117 y:6.75 z:45.55099868774414
- atom: CB x: 11.88700008392334 y:7.824999809265137 z:42.54899978637695
- atom: OG x: 12.267000198364258 y:9.003000259399414 z:43.255001068115234
- residue ALA has 5 atoms
- atom: N x: 10.75 y:6.5229997634887695 z:45.060001373291016
- atom: CA x: 10.368000030517578 y:6.559999942779541 z:46.47700119018555
- atom: C x: 11.04699993133545 y:5.427999973297119 z:47.2599983215332
- atom: O x: 11.673999786376953 y:5.670000076293945 z:48.2869987487793
- atom: CB x: 8.85200023651123 y:6.466000080108643 z:46.63600158691406
- residue ALA has 5 atoms
- atom: N x: 10.935999870300293 y:4.192999839782715 z:46.78499984741211
- atom: CA x: 11.574999809265137 y:3.0989999771118164 z:47.499000549316406
- atom: C x: 13.069999694824219 y:3.3540000915527344 z:47.66600036621094
- atom: O x: 13.619000434875488 y:3.138000011444092 z:48.733001708984375
- atom: CB x: 11.357000350952148 y:1.7940000295639038 z:46.77399826049805
- residue ARG has 11 atoms
- atom: N x: 13.737000465393066 y:3.808000087738037 z:46.617000579833984
- atom: CA x: 15.163999557495117 y:4.068999767303467 z:46.73899841308594
- atom: C x: 15.439000129699707 y:5.125999927520752 z:47.801998138427734
- atom: O x: 16.441999435424805 y:5.046999931335449 z:48.50899887084961
- atom: CB x: 15.765000343322754 y:4.5269999504089355 z:45.41400146484375
- atom: CG x: 17.187000274658203 y:5.013999938964844 z:45.55500030517578
- atom: CD x: 17.774999618530273 y:5.38100004196167 z:44.2130012512207
- atom: NE x: 17.91900062561035 y:4.206999778747559 z:43.36199951171875
- atom: CZ x: 18.624000549316406 y:3.128000020980835 z:43.6879997253418
- atom: NH1 x: 19.253999710083008 y:3.075000047683716 z:44.85200119018555
- atom: NH2 x: 18.694000244140625 y:2.1010000705718994 z:42.85100173950195
- residue LYS has 9 atoms
- atom: N x: 14.557000160217285 y:6.117000102996826 z:47.91299819946289
- atom: CA x: 14.75 y:7.156000137329102 z:48.909000396728516
- atom: C x: 14.637999534606934 y:6.564000129699707 z:50.31399917602539
- atom: O x: 15.583000183105469 y:6.656000137329102 z:51.0880012512207
- atom: CB x: 13.739999771118164 y:8.288999557495117 z:48.73099899291992
- atom: CG x: 13.96399974822998 y:9.401000022888184 z:49.72600173950195
- atom: CD x: 13.178999900817871 y:10.670000076293945 z:49.441001892089844
- atom: CE x: 13.61400032043457 y:11.75100040435791 z:50.43299865722656
- atom: NZ x: 12.967000007629395 y:13.07800006866455 z:50.22100067138672
- residue PHE has 11 atoms
- atom: N x: 13.501999855041504 y:5.952000141143799 z:50.64099884033203
- atom: CA x: 13.32699966430664 y:5.343999862670898 z:51.95600128173828
- atom: C x: 14.543999671936035 y:4.506999969482422 z:52.38100051879883
- atom: O x: 15.128999710083008 y:4.742000102996826 z:53.430999755859375
- atom: CB x: 12.090999603271484 y:4.448999881744385 z:51.974998474121094
- atom: CG x: 10.807000160217285 y:5.189000129699707 z:51.819000244140625
- atom: CD1 x: 10.470999717712402 y:6.210000038146973 z:52.6879997253418
- atom: CD2 x: 9.913000106811523 y:4.8480000495910645 z:50.8129997253418
- atom: CE1 x: 9.260000228881836 y:6.885000228881836 z:52.5629997253418
- atom: CE2 x: 8.70199966430664 y:5.513000011444092 z:50.67900085449219
- atom: CZ x: 8.378000259399414 y:6.534999847412109 z:51.560001373291016
- residue VAL has 7 atoms
- atom: N x: 14.911999702453613 y:3.5230000019073486 z:51.564998626708984
- atom: CA x: 16.048999786376953 y:2.6570000648498535 z:51.86399841308594
- atom: C x: 17.27199935913086 y:3.493000030517578 z:52.22100067138672
- atom: O x: 17.93000030517578 y:3.2239999771118164 z:53.21799850463867
- atom: CB x: 16.365999221801758 y:1.7050000429153442 z:50.66899871826172
- atom: CG1 x: 17.589000701904297 y:0.8560000061988831 z:50.96099853515625
- atom: CG2 x: 15.1850004196167 y:0.796999990940094 z:50.41600036621094
- residue ASP has 8 atoms
- atom: N x: 17.565000534057617 y:4.51800012588501 z:51.428001403808594
- atom: CA x: 18.70800018310547 y:5.386000156402588 z:51.70800018310547
- atom: C x: 18.582000732421875 y:6.093999862670898 z:53.05699920654297
- atom: O x: 19.562000274658203 y:6.625999927520752 z:53.564998626708984
- atom: CB x: 18.885000228881836 y:6.446000099182129 z:50.612998962402344
- atom: CG x: 19.240999221801758 y:5.841000080108643 z:49.27000045776367
- atom: OD1 x: 19.964000701904297 y:4.815999984741211 z:49.257999420166016
- atom: OD2 x: 18.80900001525879 y:6.396999835968018 z:48.229000091552734
- residue GLU has 9 atoms
- atom: N x: 17.381000518798828 y:6.117000102996826 z:53.62699890136719
- atom: CA x: 17.163999557495117 y:6.752999782562256 z:54.92300033569336
- atom: C x: 16.847999572753906 y:5.6570000648498535 z:55.96799850463867
- atom: O x: 16.23900032043457 y:5.9070000648498535 z:57.007999420166016
- atom: CB x: 16.024999618530273 y:7.790999889373779 z:54.821998596191406
- atom: CG x: 16.170000076293945 y:8.746000289916992 z:53.61800003051758
- atom: CD x: 15.095000267028809 y:9.843000411987305 z:53.54600143432617
- atom: OE1 x: 13.904000282287598 y:9.550999641418457 z:53.82699966430664
- atom: OE2 x: 15.446000099182129 y:10.996000289916992 z:53.183998107910156
- residue GLY has 4 atoms
- atom: N x: 17.257999420166016 y:4.429999828338623 z:55.65800094604492
- atom: CA x: 17.05900001525879 y:3.325000047683716 z:56.571998596191406
- atom: C x: 15.710000038146973 y:2.6449999809265137 z:56.645999908447266
- atom: O x: 15.593000411987305 y:1.6239999532699585 z:57.33100128173828
- residue ILE has 8 atoms
- atom: N x: 14.696999549865723 y:3.197999954223633 z:55.98099899291992
- atom: CA x: 13.359000205993652 y:2.5959999561309814 z:55.972999572753906
- atom: C x: 13.357000350952148 y:1.5210000276565552 z:54.89799880981445
- atom: O x: 13.423999786376953 y:1.8480000495910645 z:53.7130012512207
- atom: CB x: 12.27299976348877 y:3.614000082015991 z:55.59700012207031
- atom: CG1 x: 12.300999641418457 y:4.796999931335449 z:56.5620002746582
- atom: CG2 x: 10.91100025177002 y:2.947999954223633 z:55.63399887084961
- atom: CD1 x: 11.234000205993652 y:5.835999965667725 z:56.28099822998047
- residue LYS has 9 atoms
- atom: N x: 13.265000343322754 y:0.25099998712539673 z:55.29199981689453
- atom: CA x: 13.29800033569336 y:-0.828000009059906 z:54.305999755859375
- atom: C x: 12.36299991607666 y:-2.01200008392334 z:54.54800033569336
- atom: O x: 12.54699993133545 y:-3.068000078201294 z:53.96500015258789
- atom: CB x: 14.72599983215332 y:-1.3580000400543213 z:54.194000244140625
- atom: CG x: 15.210000038146973 y:-2.0199999809265137 z:55.479000091552734
- atom: CD x: 16.617000579833984 y:-2.6059999465942383 z:55.35300064086914
- atom: CE x: 17.683000564575195 y:-1.5269999504089355 z:55.19200134277344
- atom: NZ x: 19.045000076293945 y:-2.1089999675750732 z:55.125999450683594
- residue THR has 7 atoms
- atom: N x: 11.362000465393066 y:-1.8580000400543213 z:55.39500045776367
- atom: CA x: 10.472999572753906 y:-2.9719998836517334 z:55.65399932861328
- atom: C x: 9.10200023651123 y:-2.507999897003174 z:56.108001708984375
- atom: O x: 8.946999549865723 y:-1.378999948501587 z:56.555999755859375
- atom: CB x: 11.092000007629395 y:-3.8940000534057617 z:56.70600128173828
- atom: OG1 x: 11.718000411987305 y:-3.0959999561309814 z:57.71200180053711
- atom: CG2 x: 12.14799976348877 y:-4.789000034332275 z:56.07899856567383
- residue LEU has 8 atoms
- atom: N x: 8.11299991607666 y:-3.38700008392334 z:55.98899841308594
- atom: CA x: 6.741000175476074 y:-3.065000057220459 z:56.361000061035156
- atom: C x: 6.663000106811523 y:-2.3540000915527344 z:57.698001861572266
- atom: O x: 5.9079999923706055 y:-1.3930000066757202 z:57.867000579833984
- atom: CB x: 5.900000095367432 y:-4.335999965667725 z:56.41899871826172
- atom: CG x: 4.75600004196167 y:-4.4770002365112305 z:55.40999984741211
- atom: CD1 x: 5.265999794006348 y:-4.355999946594238 z:53.98500061035156
- atom: CD2 x: 4.0980000495910645 y:-5.828000068664551 z:55.604000091552734
- residue GLU has 9 atoms
- atom: N x: 7.443999767303467 y:-2.8399999141693115 z:58.652000427246094
- atom: CA x: 7.4720001220703125 y:-2.259000062942505 z:59.97999954223633
- atom: C x: 7.995999813079834 y:-0.824999988079071 z:59.93000030517578
- atom: O x: 7.494999885559082 y:0.04899999871850014 z:60.625
- atom: CB x: 8.338000297546387 y:-3.124000072479248 z:60.90299987792969
- atom: CG x: 7.71999979019165 y:-4.488999843597412 z:61.23899841308594
- atom: CD x: 7.673999786376953 y:-5.454999923706055 z:60.05400085449219
- atom: OE1 x: 8.753000259399414 y:-5.8520002365112305 z:59.5629997253418
- atom: OE2 x: 6.560999870300293 y:-5.826000213623047 z:59.61600112915039
- residue ASP has 8 atoms
- atom: N x: 9.005000114440918 y:-0.5849999785423279 z:59.104000091552734
- atom: CA x: 9.571999549865723 y:0.7480000257492065 z:58.9739990234375
- atom: C x: 8.531999588012695 y:1.718000054359436 z:58.41600036621094
- atom: O x: 8.362000465393066 y:2.8380000591278076 z:58.92399978637695
- atom: CB x: 10.795999526977539 y:0.6819999814033508 z:58.07600021362305
- atom: CG x: 11.92300033569336 y:-0.08399999886751175 z:58.7140007019043
- atom: OD1 x: 11.635000228881836 y:-1.0809999704360962 z:59.4010009765625
- atom: OD2 x: 13.090999603271484 y:0.3059999942779541 z:58.529998779296875
- residue LEU has 8 atoms
- atom: N x: 7.834000110626221 y:1.2890000343322754 z:57.37099838256836
- atom: CA x: 6.793000221252441 y:2.117000102996826 z:56.79399871826172
- atom: C x: 5.763000011444092 y:2.321000099182129 z:57.895999908447266
- atom: O x: 5.111000061035156 y:3.3589999675750732 z:57.95600128173828
- atom: CB x: 6.164000034332275 y:1.4190000295639038 z:55.58599853515625
- atom: CG x: 7.113999843597412 y:1.1069999933242798 z:54.428001403808594
- atom: CD1 x: 6.370999813079834 y:0.3479999899864197 z:53.361000061035156
- atom: CD2 x: 7.696000099182129 y:2.390000104904175 z:53.86399841308594
- residue ARG has 11 atoms
- atom: N x: 5.632999897003174 y:1.3179999589920044 z:58.76499938964844
- atom: CA x: 4.715000152587891 y:1.3680000305175781 z:59.90700149536133
- atom: C x: 5.168000221252441 y:2.438999891281128 z:60.90299987792969
- atom: O x: 4.3460001945495605 y:3.1530001163482666 z:61.48099899291992
- atom: CB x: 4.678999900817871 y:0.017999999225139618 z:60.619998931884766
- atom: CG x: 3.812999963760376 y:-1.0190000534057617 z:59.95800018310547
- atom: CD x: 2.361999988555908 y:-0.7979999780654907 z:60.33000183105469
- atom: NE x: 1.4429999589920044 y:-1.5540000200271606 z:59.483001708984375
- atom: CZ x: 1.4910000562667847 y:-2.869999885559082 z:59.29999923706055
- atom: NH1 x: 2.4179999828338623 y:-3.5999999046325684 z:59.909000396728516
- atom: NH2 x: 0.6159999966621399 y:-3.4560000896453857 z:58.4900016784668
- residue LYS has 9 atoms
- atom: N x: 6.4770002365112305 y:2.5450000762939453 z:61.104000091552734
- atom: CA x: 7.011000156402588 y:3.5309998989105225 z:62.02199935913086
- atom: C x: 6.9720001220703125 y:4.920000076293945 z:61.40399932861328
- atom: O x: 6.544000148773193 y:5.894000053405762 z:62.04800033569336
- atom: CB x: 8.460000038146973 y:3.2070000171661377 z:62.402000427246094
- atom: CG x: 9.081999778747559 y:4.255000114440918 z:63.327999114990234
- atom: CD x: 10.49899959564209 y:3.9030001163482666 z:63.790000915527344
- atom: CE x: 11.552000045776367 y:4.183000087738037 z:62.72600173950195
- atom: NZ x: 12.932000160217285 y:4.047999858856201 z:63.28200149536133
- residue ASN has 8 atoms
- atom: N x: 7.419000148773193 y:5.011000156402588 z:60.154998779296875
- atom: CA x: 7.459000110626221 y:6.291999816894531 z:59.483001708984375
- atom: C x: 6.232999801635742 y:6.5929999351501465 z:58.65599822998047
- atom: O x: 6.293000221252441 y:7.327000141143799 z:57.68299865722656
- atom: CB x: 8.72700023651123 y:6.385000228881836 z:58.63999938964844
- atom: CG x: 9.965999603271484 y:6.585999965667725 z:59.49399948120117
- atom: OD1 x: 11.059000015258789 y:6.823999881744385 z:58.986000061035156
- atom: ND2 x: 9.795000076293945 y:6.491000175476074 z:60.81100082397461
- residue GLU has 9 atoms
- atom: N x: 5.109000205993652 y:6.044000148773193 z:59.08000183105469
- atom: CA x: 3.8510000705718994 y:6.230999946594238 z:58.387001037597656
- atom: C x: 3.5390000343322754 y:7.684000015258789 z:58.02299880981445
- atom: O x: 3.0429999828338623 y:7.9670000076293945 z:56.933998107910156
- atom: CB x: 2.7279999256134033 y:5.677000045776367 z:59.244998931884766
- atom: CG x: 1.3849999904632568 y:5.681000232696533 z:58.58300018310547
- atom: CD x: 0.2879999876022339 y:5.394999980926514 z:59.57400131225586
- atom: OE1 x: 0.3709999918937683 y:4.359000205993652 z:60.27399826049805
- atom: OE2 x: -0.6549999713897705 y:6.210999965667725 z:59.65399932861328
- residue ASP has 8 atoms
- atom: N x: 3.8320000171661377 y:8.607999801635742 z:58.926998138427734
- atom: CA x: 3.5360000133514404 y:10.008000373840332 z:58.665000915527344
- atom: C x: 4.449999809265137 y:10.621000289916992 z:57.61399841308594
- atom: O x: 4.502999782562256 y:11.836999893188477 z:57.472999572753906
- atom: CB x: 3.615000009536743 y:10.812000274658203 z:59.96900177001953
- atom: CG x: 5.039000034332275 y:11.223999977111816 z:60.327999114990234
- atom: OD1 x: 5.9720001220703125 y:10.38599967956543 z:60.19599914550781
- atom: OD2 x: 5.216000080108643 y:12.392000198364258 z:60.75600051879883
- residue LYS has 9 atoms
- atom: N x: 5.159999847412109 y:9.774999618530273 z:56.874000549316406
- atom: CA x: 6.084000110626221 y:10.223999977111816 z:55.83000183105469
- atom: C x: 5.663000106811523 y:9.597999572753906 z:54.479000091552734
- atom: O x: 6.453000068664551 y:9.491999626159668 z:53.529998779296875
- atom: CB x: 7.515999794006348 y:9.8149995803833 z:56.21200180053711
- atom: CG x: 8.607000350952148 y:10.362000465393066 z:55.303001403808594
- atom: CD x: 9.99899959564209 y:9.871000289916992 z:55.709999084472656
- atom: CE x: 10.60200023651123 y:10.678999900817871 z:56.86199951171875
- atom: NZ x: 9.82699966430664 y:10.61400032043457 z:58.130001068115234
- residue LEU has 8 atoms
- atom: N x: 4.395999908447266 y:9.199000358581543 z:54.41299819946289
- atom: CA x: 3.8239998817443848 y:8.581000328063965 z:53.22800064086914
- atom: C x: 2.6070001125335693 y:9.369999885559082 z:52.75899887084961
- atom: O x: 1.850000023841858 y:9.902000427246094 z:53.573001861572266
- atom: CB x: 3.36299991607666 y:7.158999919891357 z:53.55099868774414
- atom: CG x: 4.328999996185303 y:6.171999931335449 z:54.19900131225586
- atom: CD1 x: 3.621999979019165 y:4.828999996185303 z:54.38100051879883
- atom: CD2 x: 5.563000202178955 y:6.019000053405762 z:53.32899856567383
- residue ASN has 8 atoms
- atom: N x: 2.4019999504089355 y:9.430999755859375 z:51.45000076293945
- atom: CA x: 1.2419999837875366 y:10.133999824523926 z:50.926998138427734
- atom: C x: 0.014000000432133675 y:9.26099967956543 z:51.11600112915039
- atom: O x: 0.12300000339746475 y:8.086999893188477 z:51.41600036621094
- atom: CB x: 1.4190000295639038 y:10.456000328063965 z:49.446998596191406
- atom: CG x: 1.8220000267028809 y:9.25100040435791 z:48.641998291015625
- atom: OD1 x: 1.2480000257492065 y:8.178000450134277 z:48.77899932861328
- atom: ND2 x: 2.812999963760376 y:9.425999641418457 z:47.790000915527344
- residue HIS has 10 atoms
- atom: N x: -1.1540000438690186 y:9.850000381469727 z:50.94499969482422
- atom: CA x: -2.4070000648498535 y:9.142999649047852 z:51.106998443603516
- atom: C x: -2.4030001163482666 y:7.800000190734863 z:50.391998291015625
- atom: O x: -2.8350000381469727 y:6.7829999923706055 z:50.939998626708984
- atom: CB x: -3.5369999408721924 y:10.003999710083008 z:50.566001892089844
- atom: CG x: -4.88100004196167 y:9.364999771118164 z:50.6619987487793
- atom: ND1 x: -5.572999954223633 y:9.265000343322754 z:51.8489990234375
- atom: CD2 x: -5.666999816894531 y:8.79800033569336 z:49.720001220703125
- atom: CE1 x: -6.730000019073486 y:8.668000221252441 z:51.6349983215332
- atom: NE2 x: -6.811999797821045 y:8.37399959564209 z:50.35100173950195
- residue HIS has 10 atoms
- atom: N x: -1.8949999809265137 y:7.797999858856201 z:49.17100143432617
- atom: CA x: -1.8609999418258667 y:6.583000183105469 z:48.375
- atom: C x: -0.9599999785423279 y:5.51200008392334 z:48.986000061035156
- atom: O x: -1.3209999799728394 y:4.3420000076293945 z:49.013999938964844
- atom: CB x: -1.4359999895095825 y:6.927999973297119 z:46.933998107910156
- atom: CG x: -1.406000018119812 y:5.757999897003174 z:45.99700164794922
- atom: ND1 x: -1.434999942779541 y:5.908999919891357 z:44.62900161743164
- atom: CD2 x: -1.2580000162124634 y:4.429999828338623 z:46.2239990234375
- atom: CE1 x: -1.3009999990463257 y:4.728000164031982 z:44.05400085449219
- atom: NE2 x: -1.190999984741211 y:3.815000057220459 z:45.000999450683594
- residue GLN has 9 atoms
- atom: N x: 0.20800000429153442 y:5.910999774932861 z:49.47100067138672
- atom: CA x: 1.1390000581741333 y:4.968999862670898 z:50.07500076293945
- atom: C x: 0.5680000185966492 y:4.394000053405762 z:51.38800048828125
- atom: O x: 0.800000011920929 y:3.2219998836517334 z:51.69599914550781
- atom: CB x: 2.499000072479248 y:5.65500020980835 z:50.30099868774414
- atom: CG x: 3.200000047683716 y:6.076000213623047 z:49.00299835205078
- atom: CD x: 4.383999824523926 y:7.015999794006348 z:49.22100067138672
- atom: OE1 x: 4.260000228881836 y:8.043000221252441 z:49.88100051879883
- atom: NE2 x: 5.531000137329102 y:6.669000148773193 z:48.65399932861328
- residue ARG has 11 atoms
- atom: N x: -0.18000000715255737 y:5.201000213623047 z:52.150001525878906
- atom: CA x: -0.777999997138977 y:4.710000038146973 z:53.39099884033203
- atom: C x: -1.7100000381469727 y:3.572999954223633 z:53.02399826049805
- atom: O x: -1.562999963760376 y:2.4649999141693115 z:53.52199935913086
- atom: CB x: -1.6130000352859497 y:5.7779998779296875 z:54.10200119018555
- atom: CG x: -0.9010000228881836 y:6.5980000495910645 z:55.172000885009766
- atom: CD x: -0.2150000035762787 y:7.790999889373779 z:54.56999969482422
- atom: NE x: 0.1120000034570694 y:8.810999870300293 z:55.56100082397461
- atom: CZ x: -0.7559999823570251 y:9.329000473022461 z:56.428001403808594
- atom: NH1 x: -2.0209999084472656 y:8.916000366210938 z:56.444000244140625
- atom: NH2 x: -0.3630000054836273 y:10.286999702453613 z:57.26100158691406
- residue ILE has 8 atoms
- atom: N x: -2.6760001182556152 y:3.8610000610351562 z:52.15399932861328
- atom: CA x: -3.6410000324249268 y:2.8570001125335693 z:51.694000244140625
- atom: C x: -2.88700008392334 y:1.61899995803833 z:51.20600128173828
- atom: O x: -3.2730000019073486 y:0.48100000619888306 z:51.500999450683594
- atom: CB x: -4.520999908447266 y:3.4000000953674316 z:50.507999420166016
- atom: CG1 x: -5.324999809265137 y:4.625999927520752 z:50.95399856567383
- atom: CG2 x: -5.440999984741211 y:2.302999973297119 z:49.98400115966797
- atom: CD1 x: -6.333000183105469 y:4.361000061035156 z:52.060001373291016
- residue GLY has 4 atoms
- atom: N x: -1.8070000410079956 y:1.8609999418258667 z:50.465999603271484
- atom: CA x: -0.996999979019165 y:0.7850000262260437 z:49.92599868774414
- atom: C x: -0.5400000214576721 y:-0.16500000655651093 z:51.00400161743164
- atom: O x: -0.7350000143051147 y:-1.371000051498413 z:50.88600158691406
- residue LEU has 8 atoms
- atom: N x: 0.05900000035762787 y:0.4000000059604645 z:52.053001403808594
- atom: CA x: 0.5699999928474426 y:-0.34200000762939453 z:53.20199966430664
- atom: C x: -0.5519999861717224 y:-1.0260000228881836 z:53.93899917602539
- atom: O x: -0.4169999957084656 y:-2.1730000972747803 z:54.332000732421875
- atom: CB x: 1.2879999876022339 y:0.6060000061988831 z:54.16699981689453
- atom: CG x: 1.7599999904632568 y:0.08100000023841858 z:55.53300094604492
- atom: CD1 x: 2.7009999752044678 y:-1.0989999771118164 z:55.36000061035156
- atom: CD2 x: 2.4820001125335693 y:1.2120000123977661 z:56.27399826049805
- residue LYS has 9 atoms
- atom: N x: -1.6549999713897705 y:-0.3050000071525574 z:54.11899948120117
- atom: CA x: -2.8510000705718994 y:-0.7960000038146973 z:54.805999755859375
- atom: C x: -3.437999963760376 y:-2.068000078201294 z:54.21099853515625
- atom: O x: -3.943000078201294 y:-2.9210000038146973 z:54.93199920654297
- atom: CB x: -3.940000057220459 y:0.2750000059604645 z:54.79199981689453
- atom: CG x: -5.296000003814697 y:-0.2150000035762787 z:55.270999908447266
- atom: CD x: -6.254000186920166 y:0.9430000185966492 z:55.45000076293945
- atom: CE x: -7.507999897003174 y:0.5070000290870667 z:56.176998138427734
- atom: NZ x: -8.45300006866455 y:1.6299999952316284 z:56.4119987487793
- residue TYR has 12 atoms
- atom: N x: -3.3940000534057617 y:-2.177999973297119 z:52.891998291015625
- atom: CA x: -3.928999900817871 y:-3.3459999561309814 z:52.2140007019043
- atom: C x: -2.806999921798706 y:-4.0879998207092285 z:51.51499938964844
- atom: O x: -3.015000104904175 y:-4.678999900817871 z:50.43899917602539
- atom: CB x: -4.980999946594238 y:-2.921999931335449 z:51.1870002746582
- atom: CG x: -6.118000030517578 y:-2.1530001163482666 z:51.79399871826172
- atom: CD1 x: -6.923999786376953 y:-2.7249999046325684 z:52.77000045776367
- atom: CD2 x: -6.373000144958496 y:-0.8399999737739563 z:51.41600036621094
- atom: CE1 x: -7.953999996185303 y:-2.003000020980835 z:53.3650016784668
- atom: CE2 x: -7.40500020980835 y:-0.10899999737739563 z:51.999000549316406
- atom: CZ x: -8.187999725341797 y:-0.6949999928474426 z:52.97100067138672
- atom: OH x: -9.218999862670898 y:0.01899999938905239 z:53.53499984741211
- residue PHE has 11 atoms
- atom: N x: -1.6200000047683716 y:-4.072000026702881 z:52.117000579833984
- atom: CA x: -0.4959999918937683 y:-4.738999843597412 z:51.47700119018555
- atom: C x: -0.7870000004768372 y:-6.201000213623047 z:51.15299987792969
- atom: O x: -0.6729999780654907 y:-6.626999855041504 z:50.005001068115234
- atom: CB x: 0.7450000047683716 y:-4.664000034332275 z:52.33300018310547
- atom: CG x: 1.934000015258789 y:-5.28000020980835 z:51.691001892089844
- atom: CD1 x: 2.6610000133514404 y:-4.579999923706055 z:50.74100112915039
- atom: CD2 x: 2.321000099182129 y:-6.566999912261963 z:52.01900100708008
- atom: CE1 x: 3.7679998874664307 y:-5.1479997634887695 z:50.12099838256836
- atom: CE2 x: 3.427000045776367 y:-7.1579999923706055 z:51.409000396728516
- atom: CZ x: 4.156000137329102 y:-6.446000099182129 z:50.45500183105469
- residue GLY has 4 atoms
- atom: N x: -1.159999966621399 y:-6.968999862670898 z:52.16699981689453
- atom: CA x: -1.4630000591278076 y:-8.36299991607666 z:51.941001892089844
- atom: C x: -2.6440000534057617 y:-8.527000427246094 z:51.00699996948242
- atom: O x: -2.572000026702881 y:-9.300000190734863 z:50.05500030517578
- residue ASP has 8 atoms
- atom: N x: -3.7300000190734863 y:-7.806000232696533 z:51.2760009765625
- atom: CA x: -4.914000034332275 y:-7.888999938964844 z:50.43600082397461
- atom: C x: -4.5960001945495605 y:-7.710999965667725 z:48.95600128173828
- atom: O x: -4.974999904632568 y:-8.550000190734863 z:48.137001037597656
- atom: CB x: -5.949999809265137 y:-6.8480000495910645 z:50.856998443603516
- atom: CG x: -6.870999813079834 y:-7.361999988555908 z:51.93000030517578
- atom: OD1 x: -6.843999862670898 y:-8.590999603271484 z:52.15800094604492
- atom: OD2 x: -7.624000072479248 y:-6.559999942779541 z:52.5369987487793
- residue PHE has 11 atoms
- atom: N x: -3.8889999389648438 y:-6.633999824523926 z:48.61899948120117
- atom: CA x: -3.5399999618530273 y:-6.348999977111816 z:47.233001708984375
- atom: C x: -2.683000087738037 y:-7.379000186920166 z:46.516998291015625
- atom: O x: -2.6549999713897705 y:-7.396999835968018 z:45.290000915527344
- atom: CB x: -2.8589999675750732 y:-4.974999904632568 z:47.108001708984375
- atom: CG x: -3.7939999103546143 y:-3.812000036239624 z:47.305999755859375
- atom: CD1 x: -5.133999824523926 y:-4.020999908447266 z:47.62699890136719
- atom: CD2 x: -3.3239998817443848 y:-2.5139999389648438 z:47.233001708984375
- atom: CE1 x: -5.98199987411499 y:-2.9639999866485596 z:47.88100051879883
- atom: CE2 x: -4.168000221252441 y:-1.437999963760376 z:47.48699951171875
- atom: CZ x: -5.502999782562256 y:-1.6649999618530273 z:47.814998626708984
- residue GLU has 9 atoms
- atom: N x: -1.9630000591278076 y:-8.22700023651123 z:47.244998931884766
- atom: CA x: -1.1390000581741333 y:-9.21399974822998 z:46.55099868774414
- atom: C x: -1.9110000133514404 y:-10.48900032043457 z:46.32899856567383
- atom: O x: -1.409000039100647 y:-11.414999961853027 z:45.70899963378906
- atom: CB x: 0.16899999976158142 y:-9.505000114440918 z:47.30400085449219
- atom: CG x: 1.1510000228881836 y:-8.329999923706055 z:47.29899978637695
- atom: CD x: 2.6040000915527344 y:-8.77299976348877 z:47.356998443603516
- atom: OE1 x: 2.9189999103546143 y:-9.692000389099121 z:48.141998291015625
- atom: OE2 x: 3.434000015258789 y:-8.196000099182129 z:46.624000549316406
- residue LYS has 9 atoms
- atom: N x: -3.138000011444092 y:-10.534000396728516 z:46.82699966430664
- atom: CA x: -3.9719998836517334 y:-11.711000442504883 z:46.63999938964844
- atom: C x: -4.692999839782715 y:-11.647000312805176 z:45.2859992980957
- atom: O x: -5.2129998207092285 y:-10.595000267028809 z:44.89500045776367
- atom: CB x: -5.033999919891357 y:-11.817000389099121 z:47.742000579833984
- atom: CG x: -4.519999980926514 y:-12.019000053405762 z:49.15399932861328
- atom: CD x: -5.681000232696533 y:-12.40999984741211 z:50.0629997253418
- atom: CE x: -5.315000057220459 y:-12.395000457763672 z:51.53200149536133
- atom: NZ x: -4.199999809265137 y:-13.324000358581543 z:51.86800003051758
- residue ARG has 11 atoms
- atom: N x: -4.708000183105469 y:-12.767999649047852 z:44.57099914550781
- atom: CA x: -5.4120001792907715 y:-12.85200023651123 z:43.29800033569336
- atom: C x: -6.906000137329102 y:-12.857999801635742 z:43.64899826049805
- atom: O x: -7.281000137329102 y:-13.098999977111816 z:44.79600143432617
- atom: CB x: -5.052000045776367 y:-14.151000022888184 z:42.57400131225586
- atom: CG x: -3.5899999141693115 y:-14.277000427246094 z:42.189998626708984
- atom: CD x: -3.253000020980835 y:-13.343000411987305 z:41.04800033569336
- atom: NE x: -4.007999897003174 y:-13.678000450134277 z:39.84000015258789
- atom: CZ x: -4.105000019073486 y:-12.894000053405762 z:38.768001556396484
- atom: NH1 x: -3.496999979019165 y:-11.711999893188477 z:38.73899841308594
- atom: NH2 x: -4.809000015258789 y:-13.298999786376953 z:37.71799850463867
- residue ILE has 8 atoms
- atom: N x: -7.75600004196167 y:-12.595000267028809 z:42.66600036621094
- atom: CA x: -9.199000358581543 y:-12.581000328063965 z:42.89099884033203
- atom: C x: -9.857999801635742 y:-13.762999534606934 z:42.17300033569336
- atom: O x: -9.720000267028809 y:-13.904000282287598 z:40.957000732421875
- atom: CB x: -9.817000389099121 y:-11.27400016784668 z:42.358001708984375
- atom: CG1 x: -9.052000045776367 y:-10.076000213623047 z:42.92499923706055
- atom: CG2 x: -11.300999641418457 y:-11.206000328063965 z:42.729000091552734
- atom: CD1 x: -9.505000114440918 y:-8.73900032043457 z:42.39799880981445
- residue PRO has 7 atoms
- atom: N x: -10.585000038146973 y:-14.626999855041504 z:42.9119987487793
- atom: CA x: -11.241999626159668 y:-15.781999588012695 z:42.277000427246094
- atom: C x: -12.177000045776367 y:-15.265999794006348 z:41.198001861572266
- atom: O x: -12.9399995803833 y:-14.333000183105469 z:41.44900131225586
- atom: CB x: -12.015000343322754 y:-16.426000595092773 z:43.42599868774414
- atom: CG x: -11.246999740600586 y:-15.991999626159668 z:44.65399932861328
- atom: CD x: -10.90999984741211 y:-14.5600004196167 z:44.3489990234375
- residue ARG has 11 atoms
- atom: N x: -12.13700008392334 y:-15.86400032043457 z:40.007999420166016
- atom: CA x: -12.998000144958496 y:-15.402000427246094 z:38.92300033569336
- atom: C x: -14.454999923706055 y:-15.329000473022461 z:39.33000183105469
- atom: O x: -15.230999946594238 y:-14.598999977111816 z:38.73400115966797
- atom: CB x: -12.876999855041504 y:-16.285999298095703 z:37.683998107910156
- atom: CG x: -13.494000434875488 y:-15.621000289916992 z:36.470001220703125
- atom: CD x: -13.788999557495117 y:-16.57900047302246 z:35.345001220703125
- atom: NE x: -14.845000267028809 y:-16.03700065612793 z:34.49399948120117
- atom: CZ x: -14.718000411987305 y:-14.97599983215332 z:33.696998596191406
- atom: NH1 x: -13.567000389099121 y:-14.322999954223633 z:33.61399841308594
- atom: NH2 x: -15.767000198364258 y:-14.541999816894531 z:33.007999420166016
- residue GLU has 9 atoms
- atom: N x: -14.829000473022461 y:-16.084999084472656 z:40.34700012207031
- atom: CA x: -16.200000762939453 y:-16.082000732421875 z:40.83100128173828
- atom: C x: -16.530000686645508 y:-14.770999908447266 z:41.529998779296875
- atom: O x: -17.562000274658203 y:-14.151000022888184 z:41.26499938964844
- atom: CB x: -16.413999557495117 y:-17.25200080871582 z:41.79399871826172
- atom: CG x: -17.667999267578125 y:-17.152000427246094 z:42.63999938964844
- atom: CD x: -18.077999114990234 y:-18.496999740600586 z:43.21699905395508
- atom: OE1 x: -18.999000549316406 y:-18.52400016784668 z:44.064998626708984
- atom: OE2 x: -17.48699951171875 y:-19.530000686645508 z:42.816001892089844
- residue GLU has 9 atoms
- atom: N x: -15.652999877929688 y:-14.359000205993652 z:42.4370002746582
- atom: CA x: -15.861000061035156 y:-13.123000144958496 z:43.167999267578125
- atom: C x: -15.913000106811523 y:-11.949999809265137 z:42.1879997253418
- atom: O x: -16.642000198364258 y:-10.979999542236328 z:42.409000396728516
- atom: CB x: -14.75100040435791 y:-12.937999725341797 z:44.20800018310547
- atom: CG x: -14.975000381469727 y:-13.76099967956543 z:45.46699905395508
- atom: CD x: -13.857000350952148 y:-13.616000175476074 z:46.49100112915039
- atom: OE1 x: -13.395000457763672 y:-12.477999687194824 z:46.71699905395508
- atom: OE2 x: -13.449999809265137 y:-14.640999794006348 z:47.08100128173828
- residue MET has 8 atoms
- atom: N x: -15.138999938964844 y:-12.048999786376953 z:41.106998443603516
- atom: CA x: -15.130000114440918 y:-11.010000228881836 z:40.0890007019043
- atom: C x: -16.57699966430664 y:-10.881999969482422 z:39.64799880981445
- atom: O x: -17.172000885009766 y:-9.8100004196167 z:39.71099853515625
- atom: CB x: -14.267999649047852 y:-11.425999641418457 z:38.893001556396484
- atom: CG x: -12.784000396728516 y:-11.329000473022461 z:39.141998291015625
- atom: SD x: -12.277000427246094 y:-9.61400032043457 z:39.38199996948242
- atom: CE x: -11.970999717712402 y:-9.13599967956543 z:37.689998626708984
- residue LEU has 8 atoms
- atom: N x: -17.142000198364258 y:-12.005999565124512 z:39.222999572753906
- atom: CA x: -18.52199935913086 y:-12.043999671936035 z:38.777000427246094
- atom: C x: -19.43400001525879 y:-11.295000076293945 z:39.74399948120117
- atom: O x: -20.256000518798828 y:-10.489999771118164 z:39.3129997253418
- atom: CB x: -18.979000091552734 y:-13.49899959564209 z:38.625999450683594
- atom: CG x: -18.22599983215332 y:-14.336000442504883 z:37.57699966430664
- atom: CD1 x: -18.56999969482422 y:-15.805999755859375 z:37.749000549316406
- atom: CD2 x: -18.573999404907227 y:-13.864999771118164 z:36.16899871826172
- residue GLN has 9 atoms
- atom: N x: -19.2810001373291 y:-11.53499984741211 z:41.04499816894531
- atom: CA x: -20.1299991607666 y:-10.86299991607666 z:42.040000915527344
- atom: C x: -19.868999481201172 y:-9.359000205993652 z:42.04899978637695
- atom: O x: -20.73900032043457 y:-8.557000160217285 z:42.40599822998047
- atom: CB x: -19.88800048828125 y:-11.430999755859375 z:43.44499969482422
- atom: CG x: -19.590999603271484 y:-12.925000190734863 z:43.465999603271484
- atom: CD x: -19.812999725341797 y:-13.557999610900879 z:44.827999114990234
- atom: OE1 x: -19.44499969482422 y:-14.71399974822998 z:45.05099868774414
- atom: NE2 x: -20.426000595092773 y:-12.809000015258789 z:45.742000579833984
- residue MET has 8 atoms
- atom: N x: -18.6560001373291 y:-8.982999801635742 z:41.66299819946289
- atom: CA x: -18.284000396728516 y:-7.578999996185303 z:41.612998962402344
- atom: C x: -18.82900047302246 y:-7.03000020980835 z:40.29999923706055
- atom: O x: -19.385000228881836 y:-5.929999828338623 z:40.25899887084961
- atom: CB x: -16.759000778198242 y:-7.435999870300293 z:41.68299865722656
- atom: CG x: -16.158000946044922 y:-7.941999912261963 z:42.99399948120117
- atom: SD x: -14.380999565124512 y:-8.246999740600586 z:42.926998138427734
- atom: CE x: -13.717000007629395 y:-6.666999816894531 z:43.327999114990234
- residue GLN has 9 atoms
- atom: N x: -18.690000534057617 y:-7.821000099182129 z:39.236000061035156
- atom: CA x: -19.16900062561035 y:-7.436999797821045 z:37.9119987487793
- atom: C x: -20.66699981689453 y:-7.296000003814697 z:37.957000732421875
- atom: O x: -21.280000686645508 y:-6.793000221252441 z:37.0260009765625
- atom: CB x: -18.819000244140625 y:-8.5 z:36.875999450683594
- atom: CG x: -19.424999237060547 y:-8.256999969482422 z:35.5
- atom: CD x: -19.215999603271484 y:-9.430999755859375 z:34.56100082397461
- atom: OE1 x: -19.83799934387207 y:-10.493000030517578 z:34.7140007019043
- atom: NE2 x: -18.32699966430664 y:-9.251999855041504 z:33.58399963378906
- residue ASP has 8 atoms
- atom: N x: -21.257999420166016 y:-7.749000072479248 z:39.04999923706055
- atom: CA x: -22.69499969482422 y:-7.681000232696533 z:39.194000244140625
- atom: C x: -23.049999237060547 y:-6.394999980926514 z:39.89699935913086
- atom: O x: -23.94499969482422 y:-5.671999931335449 z:39.46900177001953
- atom: CB x: -23.18600082397461 y:-8.876999855041504 z:40.00299835205078
- atom: CG x: -24.58799934387207 y:-9.300999641418457 z:39.630001068115234
- atom: OD1 x: -25.54599952697754 y:-8.54699993133545 z:39.91600036621094
- atom: OD2 x: -24.722000122070312 y:-10.397000312805176 z:39.04600143432617
- residue ILE has 8 atoms
- atom: N x: -22.33099937438965 y:-6.107999801635742 z:40.97600173950195
- atom: CA x: -22.56999969482422 y:-4.901000022888184 z:41.7599983215332
- atom: C x: -22.198999404907227 y:-3.635999917984009 z:40.992000579833984
- atom: O x: -22.82900047302246 y:-2.5940001010894775 z:41.16600036621094
- atom: CB x: -21.783000946044922 y:-4.942999839782715 z:43.0989990234375
- atom: CG1 x: -22.229999542236328 y:-6.1579999923706055 z:43.92100143432617
- atom: CG2 x: -22.006999969482422 y:-3.6570000648498535 z:43.88999938964844
- atom: CD1 x: -21.645999908447266 y:-6.208000183105469 z:45.327999114990234
- residue VAL has 7 atoms
- atom: N x: -21.18000030517578 y:-3.7290000915527344 z:40.14099884033203
- atom: CA x: -20.750999450683594 y:-2.5759999752044678 z:39.361000061035156
- atom: C x: -21.81100082397461 y:-2.3359999656677246 z:38.30500030517578
- atom: O x: -22.53499984741211 y:-1.3489999771118164 z:38.35900115966797
- atom: CB x: -19.393999099731445 y:-2.825000047683716 z:38.67300033569336
- atom: CG1 x: -18.95199966430664 y:-1.5759999752044678 z:37.946998596191406
- atom: CG2 x: -18.347000122070312 y:-3.2209999561309814 z:39.69900131225586
- residue LEU has 8 atoms
- atom: N x: -21.909000396728516 y:-3.263000011444092 z:37.358001708984375
- atom: CA x: -22.89699935913086 y:-3.177999973297119 z:36.290000915527344
- atom: C x: -24.26300048828125 y:-2.7330000400543213 z:36.8120002746582
- atom: O x: -24.941999435424805 y:-1.906000018119812 z:36.19200134277344
- atom: CB x: -23.040000915527344 y:-4.533999919891357 z:35.59600067138672
- atom: CG x: -22.295000076293945 y:-4.75 z:34.27399826049805
- atom: CD1 x: -20.80900001525879 y:-4.521999835968018 z:34.46500015258789
- atom: CD2 x: -22.566999435424805 y:-6.160999774932861 z:33.75899887084961
- residue ASN has 8 atoms
- atom: N x: -24.6560001373291 y:-3.2720000743865967 z:37.96200180053711
- atom: CA x: -25.947999954223633 y:-2.943000078201294 z:38.551998138427734
- atom: C x: -26.011999130249023 y:-1.4980000257492065 z:39.018001556396484
- atom: O x: -26.774999618530273 y:-0.6959999799728394 z:38.49100112915039
- atom: CB x: -26.240999221801758 y:-3.865999937057495 z:39.736000061035156
- atom: CG x: -27.707000732421875 y:-3.8570001125335693 z:40.130001068115234
- atom: OD1 x: -28.28499984741211 y:-2.805999994277954 z:40.42100143432617
- atom: ND2 x: -28.31800079345703 y:-5.035999774932861 z:40.14099884033203
- residue GLU has 9 atoms
- atom: N x: -25.208999633789062 y:-1.1749999523162842 z:40.02000045776367
- atom: CA x: -25.179000854492188 y:0.16699999570846558 z:40.577999114990234
- atom: C x: -25.06399917602539 y:1.25 z:39.516998291015625
- atom: O x: -25.56100082397461 y:2.3570001125335693 z:39.70199966430664
- atom: CB x: -24.020999908447266 y:0.2759999930858612 z:41.569000244140625
- atom: CG x: -24.288000106811523 y:-0.4399999976158142 z:42.861000061035156
- atom: CD x: -25.4689998626709 y:0.15700000524520874 z:43.57600021362305
- atom: OE1 x: -25.375999450683594 y:1.3259999752044678 z:43.999000549316406
- atom: OE2 x: -26.4950008392334 y:-0.5339999794960022 z:43.70399856567383
- residue VAL has 7 atoms
- atom: N x: -24.398000717163086 y:0.9309999942779541 z:38.41400146484375
- atom: CA x: -24.229999542236328 y:1.8899999856948853 z:37.33399963378906
- atom: C x: -25.618999481201172 y:2.247999906539917 z:36.79499816894531
- atom: O x: -25.95599937438965 y:3.4240000247955322 z:36.630001068115234
- atom: CB x: -23.381000518798828 y:1.2999999523162842 z:36.17900085449219
- atom: CG1 x: -23.075000762939453 y:2.36899995803833 z:35.16899871826172
- atom: CG2 x: -22.10099983215332 y:0.7269999980926514 z:36.702999114990234
- residue LYS has 9 atoms
- atom: N x: -26.413999557495117 y:1.215000033378601 z:36.52299880981445
- atom: CA x: -27.770000457763672 y:1.378000020980835 z:36.016998291015625
- atom: C x: -28.56100082397461 y:2.3529999256134033 z:36.87699890136719
- atom: O x: -29.222999572753906 y:3.249000072479248 z:36.358001708984375
- atom: CB x: -28.500999450683594 y:0.03500000014901161 z:36.00199890136719
- atom: CG x: -29.937999725341797 y:0.13699999451637268 z:35.51100158691406
- atom: CD x: -30.6560001373291 y:-1.2070000171661377 z:35.52299880981445
- atom: CE x: -31.038000106811523 y:-1.6349999904632568 z:36.933998107910156
- atom: NZ x: -31.767000198364258 y:-2.940999984741211 z:36.91899871826172
- residue LYS has 9 atoms
- atom: N x: -28.483999252319336 y:2.1700000762939453 z:38.189998626708984
- atom: CA x: -29.195999145507812 y:3.01200008392334 z:39.145999908447266
- atom: C x: -28.899999618530273 y:4.515999794006348 z:39.060001373291016
- atom: O x: -29.618000030517578 y:5.326000213623047 z:39.63399887084961
- atom: CB x: -28.916000366210938 y:2.5190000534057617 z:40.564998626708984
- atom: CG x: -29.30299949645996 y:1.0679999589920044 z:40.792999267578125
- atom: CD x: -29.030000686645508 y:0.6269999742507935 z:42.22800064086914
- atom: CE x: -29.3700008392334 y:-0.847000002861023 z:42.40999984741211
- atom: NZ x: -29.13800048828125 y:-1.3320000171661377 z:43.79899978637695
- residue VAL has 7 atoms
- atom: N x: -27.844999313354492 y:4.8979997634887695 z:38.35499954223633
- atom: CA x: -27.518999099731445 y:6.308000087738037 z:38.22700119018555
- atom: C x: -28.152000427246094 y:6.7729997634887695 z:36.92399978637695
- atom: O x: -28.701000213623047 y:7.872000217437744 z:36.827999114990234
- atom: CB x: -25.989999771118164 y:6.511000156402588 z:38.16600036621094
- atom: CG1 x: -25.635000228881836 y:7.994999885559082 z:38.32899856567383
- atom: CG2 x: -25.32699966430664 y:5.671000003814697 z:39.244998931884766
- residue ASP has 8 atoms
- atom: N x: -28.059999465942383 y:5.916999816894531 z:35.91699981689453
- atom: CA x: -28.628000259399414 y:6.202000141143799 z:34.62099838256836
- atom: C x: -28.538000106811523 y:4.939000129699707 z:33.79800033569336
- atom: O x: -27.46500015258789 y:4.364999771118164 z:33.65299987792969
- atom: CB x: -27.874000549316406 y:7.335000038146973 z:33.917999267578125
- atom: CG x: -28.533000946044922 y:7.744999885559082 z:32.60300064086914
- atom: OD1 x: -29.73699951171875 y:8.086999893188477 z:32.624000549316406
- atom: OD2 x: -27.854000091552734 y:7.7210001945495605 z:31.55299949645996
- residue SER has 6 atoms
- atom: N x: -29.67799949645996 y:4.511000156402588 z:33.266998291015625
- atom: CA x: -29.750999450683594 y:3.305000066757202 z:32.45500183105469
- atom: C x: -29.03700065612793 y:3.424999952316284 z:31.115999221801758
- atom: O x: -29.08099937438965 y:2.493000030517578 z:30.319000244140625
- atom: CB x: -31.204999923706055 y:2.9260001182556152 z:32.183998107910156
- atom: OG x: -31.709999084472656 y:3.6610000133514404 z:31.082000732421875
- residue GLU has 9 atoms
- atom: N x: -28.38800048828125 y:4.552000045776367 z:30.847999572753906
- atom: CA x: -27.687999725341797 y:4.685999870300293 z:29.575000762939453
- atom: C x: -26.202999114990234 y:4.35099983215332 z:29.645999908447266
- atom: O x: -25.54199981689453 y:4.173999786376953 z:28.621999740600586
- atom: CB x: -27.898000717163086 y:6.085999965667725 z:29.003999710083008
- atom: CG x: -29.305999755859375 y:6.269999980926514 z:28.450000762939453
- atom: CD x: -29.743999481201172 y:5.085999965667725 z:27.591999053955078
- atom: OE1 x: -29.18400001525879 y:4.901000022888184 z:26.48900032043457
- atom: OE2 x: -30.64299964904785 y:4.333000183105469 z:28.02899932861328
- residue TYR has 12 atoms
- atom: N x: -25.69300079345703 y:4.261000156402588 z:30.868000030517578
- atom: CA x: -24.302000045776367 y:3.9110000133514404 z:31.118000030517578
- atom: C x: -24.009000778198242 y:2.4860000610351562 z:30.6299991607666
- atom: O x: -24.80500030517578 y:1.5779999494552612 z:30.854000091552734
- atom: CB x: -24.033000946044922 y:3.9600000381469727 z:32.612998962402344
- atom: CG x: -23.69300079345703 y:5.311999797821045 z:33.18600082397461
- atom: CD1 x: -22.573999404907227 y:6.00600004196167 z:32.75400161743164
- atom: CD2 x: -24.45400047302246 y:5.866000175476074 z:34.209999084472656
- atom: CE1 x: -22.222999572753906 y:7.206999778747559 z:33.33100128173828
- atom: CE2 x: -24.111000061035156 y:7.065999984741211 z:34.78499984741211
- atom: CZ x: -22.993999481201172 y:7.724999904632568 z:34.340999603271484
- atom: OH x: -22.635000228881836 y:8.914999961853027 z:34.90700149536133
- residue ILE has 8 atoms
- atom: N x: -22.875 y:2.2880001068115234 z:29.96299934387207
- atom: CA x: -22.486000061035156 y:0.9539999961853027 z:29.506000518798828
- atom: C x: -21.17099952697754 y:0.5669999718666077 z:30.18600082397461
- atom: O x: -20.097000122070312 y:0.6959999799728394 z:29.594999313354492
- atom: CB x: -22.257999420166016 y:0.8889999985694885 z:27.974000930786133
- atom: CG1 x: -23.577999114990234 y:1.0160000324249268 z:27.23200035095215
- atom: CG2 x: -21.613000869750977 y:-0.45100000500679016 z:27.584999084472656
- atom: CD1 x: -23.41699981689453 y:0.8740000128746033 z:25.736000061035156
- residue ALA has 5 atoms
- atom: N x: -21.249000549316406 y:0.10000000149011612 z:31.42799949645996
- atom: CA x: -20.040000915527344 y:-0.296999990940094 z:32.15399932861328
- atom: C x: -19.613000869750977 y:-1.6979999542236328 z:31.74799919128418
- atom: O x: -20.31399917602539 y:-2.6679999828338623 z:32.02799987792969
- atom: CB x: -20.28700065612793 y:-0.24199999868869781 z:33.65700149536133
- residue THR has 7 atoms
- atom: N x: -18.457000732421875 y:-1.7899999618530273 z:31.097999572753906
- atom: CA x: -17.916000366210938 y:-3.066999912261963 z:30.64299964904785
- atom: C x: -16.632999420166016 y:-3.4679999351501465 z:31.371000289916992
- atom: O x: -15.590999603271484 y:-2.819999933242798 z:31.200000762939453
- atom: CB x: -17.61199951171875 y:-3.0190000534057617 z:29.134000778198242
- atom: OG1 x: -18.81399917602539 y:-2.7090001106262207 z:28.420000076293945
- atom: CG2 x: -17.062000274658203 y:-4.354000091552734 z:28.64299964904785
- residue VAL has 7 atoms
- atom: N x: -16.701000213623047 y:-4.531000137329102 z:32.178001403808594
- atom: CA x: -15.51200008392334 y:-5.008999824523926 z:32.893001556396484
- atom: C x: -14.501999855041504 y:-5.539999961853027 z:31.885000228881836
- atom: O x: -14.866999626159668 y:-6.291999816894531 z:31.0
- atom: CB x: -15.82800006866455 y:-6.168000221252441 z:33.861000061035156
- atom: CG1 x: -14.53499984741211 y:-6.660999774932861 z:34.49399948120117
- atom: CG2 x: -16.80299949645996 y:-5.7170000076293945 z:34.933998107910156
- residue CYS has 6 atoms
- atom: N x: -13.236000061035156 y:-5.158999919891357 z:32.01599884033203
- atom: CA x: -12.208000183105469 y:-5.639999866485596 z:31.089000701904297
- atom: C x: -10.972000122070312 y:-6.2220001220703125 z:31.773000717163086
- atom: O x: -11.039999961853027 y:-6.798999786376953 z:32.85300064086914
- atom: CB x: -11.751999855041504 y:-4.51800012588501 z:30.163000106811523
- atom: SG x: -13.08899974822998 y:-3.6470000743865967 z:29.341999053955078
- residue GLY has 4 atoms
- atom: N x: -9.829999923706055 y:-6.051000118255615 z:31.124000549316406
- atom: CA x: -8.604000091552734 y:-6.572999954223633 z:31.680999755859375
- atom: C x: -8.550999641418457 y:-8.071999549865723 z:31.476999282836914
- atom: O x: -9.340999603271484 y:-8.630000114440918 z:30.722000122070312
- residue SER has 6 atoms
- atom: N x: -7.61899995803833 y:-8.729999542236328 z:32.15399932861328
- atom: CA x: -7.484000205993652 y:-10.166000366210938 z:32.025001525878906
- atom: C x: -8.803000450134277 y:-10.85200023651123 z:32.362998962402344
- atom: O x: -8.97700023651123 y:-12.031000137329102 z:32.077999114990234
- atom: CB x: -6.353000164031982 y:-10.680999755859375 z:32.926998138427734
- atom: OG x: -6.578000068664551 y:-10.366000175476074 z:34.28799819946289
- residue PHE has 11 atoms
- atom: N x: -9.734999656677246 y:-10.123000144958496 z:32.96799850463867
- atom: CA x: -11.02400016784668 y:-10.720999717712402 z:33.28300094604492
- atom: C x: -11.821999549865723 y:-10.930999755859375 z:32.000999450683594
- atom: O x: -12.414999961853027 y:-11.984999656677246 z:31.797000885009766
- atom: CB x: -11.848999977111816 y:-9.84000015258789 z:34.20500183105469
- atom: CG x: -13.270000457763672 y:-10.277000427246094 z:34.305999755859375
- atom: CD1 x: -13.663000106811523 y:-11.180000305175781 z:35.28200149536133
- atom: CD2 x: -14.206999778747559 y:-9.845999717712402 z:33.369998931884766
- atom: CE1 x: -14.968000411987305 y:-11.656000137329102 z:35.32699966430664
- atom: CE2 x: -15.517999649047852 y:-10.312000274658203 z:33.39799880981445
- atom: CZ x: -15.902999877929688 y:-11.220999717712402 z:34.37900161743164
- residue ARG has 11 atoms
- atom: N x: -11.862000465393066 y:-9.913000106811523 z:31.148000717163086
- atom: CA x: -12.595000267028809 y:-10.017999649047852 z:29.895000457763672
- atom: C x: -11.9350004196167 y:-11.093999862670898 z:29.05299949645996
- atom: O x: -12.581999778747559 y:-11.767000198364258 z:28.249000549316406
- atom: CB x: -12.574000358581543 y:-8.678999900817871 z:29.16200065612793
- atom: CG x: -13.28499984741211 y:-8.682000160217285 z:27.829999923706055
- atom: CD x: -14.185999870300293 y:-7.460000038146973 z:27.68899917602539
- atom: NE x: -14.734000205993652 y:-7.388000011444092 z:26.347000122070312
- atom: CZ x: -13.996999740600586 y:-7.210000038146973 z:25.257999420166016
- atom: NH1 x: -12.6850004196167 y:-7.070000171661377 z:25.35300064086914
- atom: NH2 x: -14.567999839782715 y:-7.215000152587891 z:24.062999725341797
- residue ARG has 11 atoms
- atom: N x: -10.635000228881836 y:-11.258999824523926 z:29.259000778198242
- atom: CA x: -9.885000228881836 y:-12.258999824523926 z:28.534000396728516
- atom: C x: -10.24899959564209 y:-13.630999565124512 z:29.086000442504883
- atom: O x: -9.66100025177002 y:-14.633999824523926 z:28.714000701904297
- atom: CB x: -8.387999534606934 y:-12.006999969482422 z:28.687999725341797
- atom: CG x: -7.964000225067139 y:-10.605999946594238 z:28.301000595092773
- atom: CD x: -6.538000106811523 y:-10.583000183105469 z:27.792999267578125
- atom: NE x: -5.581999778747559 y:-11.0600004196167 z:28.784000396728516
- atom: CZ x: -5.169000148773193 y:-10.347000122070312 z:29.82200050354004
- atom: NH1 x: -5.623000144958496 y:-9.119000434875488 z:30.011999130249023
- atom: NH2 x: -4.296000003814697 y:-10.855999946594238 z:30.66900062561035
- residue GLY has 4 atoms
- atom: N x: -11.241999626159668 y:-13.666000366210938 z:29.965999603271484
- atom: CA x: -11.654000282287598 y:-14.925000190734863 z:30.55900001525879
- atom: C x: -10.503999710083008 y:-15.541999816894531 z:31.332000732421875
- atom: O x: -9.788000106811523 y:-16.39900016784668 z:30.81100082397461
- residue ALA has 5 atoms
- atom: N x: -10.319000244140625 y:-15.102999687194824 z:32.573001861572266
- atom: CA x: -9.23900032043457 y:-15.609000205993652 z:33.4109992980957
- atom: C x: -9.798999786376953 y:-16.110000610351562 z:34.7239990234375
- atom: O x: -10.76099967956543 y:-15.543999671936035 z:35.250999450683594
- atom: CB x: -8.211999893188477 y:-14.51099967956543 z:33.6619987487793
- residue GLU has 9 atoms
- atom: N x: -9.192999839782715 y:-17.165000915527344 z:35.25699996948242
- atom: CA x: -9.666000366210938 y:-17.74799919128418 z:36.512001037597656
- atom: C x: -9.555000305175781 y:-16.80900001525879 z:37.69599914550781
- atom: O x: -10.270000457763672 y:-16.94700050354004 z:38.68600082397461
- atom: CB x: -8.932000160217285 y:-19.05900001525879 z:36.80099868774414
- atom: CG x: -9.420000076293945 y:-20.18199920654297 z:35.91999816894531
- atom: CD x: -10.944999694824219 y:-20.224000930786133 z:35.84400177001953
- atom: OE1 x: -11.60200023651123 y:-20.437999725341797 z:36.89699935913086
- atom: OE2 x: -11.484999656677246 y:-20.034000396728516 z:34.72700119018555
- residue SER has 6 atoms
- atom: N x: -8.654999732971191 y:-15.845999717712402 z:37.58000183105469
- atom: CA x: -8.458000183105469 y:-14.861000061035156 z:38.625999450683594
- atom: C x: -7.815999984741211 y:-13.618000030517578 z:38.0099983215332
- atom: O x: -7.223999977111816 y:-13.684000015258789 z:36.92599868774414
- atom: CB x: -7.578000068664551 y:-15.440999984741211 z:39.744998931884766
- atom: OG x: -6.326000213623047 y:-15.878999710083008 z:39.25
- residue SER has 6 atoms
- atom: N x: -7.935999870300293 y:-12.489999771118164 z:38.70100021362305
- atom: CA x: -7.374000072479248 y:-11.253000259399414 z:38.196998596191406
- atom: C x: -6.690999984741211 y:-10.42300033569336 z:39.26900100708008
- atom: O x: -7.107999801635742 y:-10.406000137329102 z:40.42399978637695
- atom: CB x: -8.47599983215332 y:-10.42199993133545 z:37.54199981689453
- atom: OG x: -9.085000038146973 y:-11.130999565124512 z:36.481998443603516
- residue GLY has 4 atoms
- atom: N x: -5.633999824523926 y:-9.72700023651123 z:38.882999420166016
- atom: CA x: -4.954999923706055 y:-8.890999794006348 z:39.84299850463867
- atom: C x: -5.926000118255615 y:-7.854000091552734 z:40.37900161743164
- atom: O x: -5.889999866485596 y:-7.50600004196167 z:41.551998138427734
- residue ASP has 8 atoms
- atom: N x: -6.810999870300293 y:-7.369999885559082 z:39.51900100708008
- atom: CA x: -7.783999919891357 y:-6.349999904632568 z:39.9119987487793
- atom: C x: -9.005000114440918 y:-6.341000080108643 z:38.992000579833984
- atom: O x: -9.175000190734863 y:-7.230999946594238 z:38.16699981689453
- atom: CB x: -7.130000114440918 y:-4.9670000076293945 z:39.8650016784668
- atom: CG x: -6.76800012588501 y:-4.546000003814697 z:38.452999114990234
- atom: OD1 x: -6.9720001220703125 y:-5.339000225067139 z:37.50899887084961
- atom: OD2 x: -6.2779998779296875 y:-3.4170000553131104 z:38.27899932861328
- residue MET has 8 atoms
- atom: N x: -9.845999717712402 y:-5.321000099182129 z:39.1510009765625
- atom: CA x: -11.045000076293945 y:-5.1479997634887695 z:38.334999084472656
- atom: C x: -10.956000328063965 y:-3.8489999771118164 z:37.55500030517578
- atom: O x: -10.967000007629395 y:-2.7790000438690186 z:38.13800048828125
- atom: CB x: -12.298999786376953 y:-5.109000205993652 z:39.194000244140625
- atom: CG x: -13.512999534606934 y:-4.709000110626221 z:38.38399887084961
- atom: SD x: -15.081000328063965 y:-5.085999965667725 z:39.16600036621094
- atom: CE x: -15.109999656677246 y:-6.809000015258789 z:38.875
- residue ASP has 8 atoms
- atom: N x: -10.873000144958496 y:-3.950000047683716 z:36.23500061035156
- atom: CA x: -10.781000137329102 y:-2.7780001163482666 z:35.367000579833984
- atom: C x: -12.133000373840332 y:-2.5320000648498535 z:34.70500183105469
- atom: O x: -12.66100025177002 y:-3.4049999713897705 z:34.04100036621094
- atom: CB x: -9.710000038146973 y:-3.0380001068115234 z:34.321998596191406
- atom: CG x: -8.418999671936035 y:-3.5199999809265137 z:34.94300079345703
- atom: OD1 x: -7.6579999923706055 y:-2.6649999618530273 z:35.45100021362305
- atom: OD2 x: -8.175999641418457 y:-4.752999782562256 z:34.95899963378906
- residue VAL has 7 atoms
- atom: N x: -12.694000244140625 y:-1.3459999561309814 z:34.887001037597656
- atom: CA x: -13.998000144958496 y:-1.0369999408721924 z:34.314998626708984
- atom: C x: -13.968999862670898 y:0.0860000029206276 z:33.28300094604492
- atom: O x: -13.496000289916992 y:1.1820000410079956 z:33.5629997253418
- atom: CB x: -14.99899959564209 y:-0.6549999713897705 z:35.433998107910156
- atom: CG1 x: -16.375 y:-0.4050000011920929 z:34.85599899291992
- atom: CG2 x: -15.0649995803833 y:-1.7660000324249268 z:36.45800018310547
- residue LEU has 8 atoms
- atom: N x: -14.458000183105469 y:-0.19699999690055847 z:32.082000732421875
- atom: CA x: -14.527999877929688 y:0.8050000071525574 z:31.0310001373291
- atom: C x: -15.949999809265137 y:1.3270000219345093 z:31.106000900268555
- atom: O x: -16.90399932861328 y:0.5569999814033508 z:31.158000946044922
- atom: CB x: -14.281000137329102 y:0.19300000369548798 z:29.663999557495117
- atom: CG x: -14.069000244140625 y:1.2020000219345093 z:28.530000686645508
- atom: CD1 x: -12.979000091552734 y:2.184999942779541 z:28.922000885009766
- atom: CD2 x: -13.649999618530273 y:0.48399999737739563 z:27.270000457763672
- residue LEU has 8 atoms
- atom: N x: -16.086999893188477 y:2.640000104904175 z:31.107999801635742
- atom: CA x: -17.38800048828125 y:3.260999917984009 z:31.239999771118164
- atom: C x: -17.714000701904297 y:4.26200008392334 z:30.121999740600586
- atom: O x: -16.847000122070312 y:5.017000198364258 z:29.67799949645996
- atom: CB x: -17.423999786376953 y:3.9570000171661377 z:32.59700012207031
- atom: CG x: -18.70199966430664 y:4.685999870300293 z:32.948001861572266
- atom: CD1 x: -19.889999389648438 y:3.7049999237060547 z:32.92599868774414
- atom: CD2 x: -18.520000457763672 y:5.311999797821045 z:34.292999267578125
- residue THR has 7 atoms
- atom: N x: -18.966999053955078 y:4.270999908447266 z:29.67099952697754
- atom: CA x: -19.382999420166016 y:5.206999778747559 z:28.621999740600586
- atom: C x: -20.781999588012695 y:5.7170000076293945 z:28.87299919128418
- atom: O x: -21.530000686645508 y:5.166999816894531 z:29.683000564575195
- atom: CB x: -19.409000396728516 y:4.563000202178955 z:27.23900032043457
- atom: OG1 x: -20.46500015258789 y:3.6029999256134033 z:27.19499969482422
- atom: CG2 x: -18.097000122070312 y:3.871000051498413 z:26.937999725341797
- residue HIS has 10 atoms
- atom: N x: -21.13599967956543 y:6.7769999504089355 z:28.163999557495117
- atom: CA x: -22.46500015258789 y:7.359000205993652 z:28.28700065612793
- atom: C x: -22.768999099731445 y:8.1899995803833 z:27.04800033569336
- atom: O x: -21.88599967956543 y:8.836999893188477 z:26.488000869750977
- atom: CB x: -22.566999435424805 y:8.227999687194824 z:29.542999267578125
- atom: CG x: -23.91200065612793 y:8.859999656677246 z:29.735000610351562
- atom: ND1 x: -24.489999771118164 y:9.684000015258789 z:28.79199981689453
- atom: CD2 x: -24.777999877929688 y:8.817000389099121 z:30.777000427246094
- atom: CE1 x: -25.650999069213867 y:10.123000144958496 z:29.243999481201172
- atom: NE2 x: -25.850000381469727 y:9.612000465393066 z:30.44700050354004
- residue PRO has 7 atoms
- atom: N x: -24.027999877929688 y:8.15999984741211 z:26.590999603271484
- atom: CA x: -24.441999435424805 y:8.918999671936035 z:25.409000396728516
- atom: C x: -24.042999267578125 y:10.402999877929688 z:25.429000854492188
- atom: O x: -23.601999282836914 y:10.946000099182129 z:24.416000366210938
- atom: CB x: -25.95199966430664 y:8.711999893188477 z:25.395999908447266
- atom: CG x: -26.070999145507812 y:7.288000106811523 z:25.88599967956543
- atom: CD x: -25.117000579833984 y:7.2789998054504395 z:27.059999465942383
- residue SER has 6 atoms
- atom: N x: -24.176000595092773 y:11.045000076293945 z:26.583999633789062
- atom: CA x: -23.854000091552734 y:12.461999893188477 z:26.711000442504883
- atom: C x: -22.368000030517578 y:12.763999938964844 z:26.673999786376953
- atom: O x: -21.957000732421875 y:13.888999938964844 z:26.972999572753906
- atom: CB x: -24.410999298095703 y:13.015000343322754 z:28.018999099731445
- atom: OG x: -23.520000457763672 y:12.762999534606934 z:29.091999053955078
- residue PHE has 11 atoms
- atom: N x: -21.558000564575195 y:11.779999732971191 z:26.30699920654297
- atom: CA x: -20.12299919128418 y:11.99899959564209 z:26.285999298095703
- atom: C x: -19.398000717163086 y:11.35200023651123 z:25.132999420166016
- atom: O x: -19.242000579833984 y:10.137999534606934 z:25.084999084472656
- atom: CB x: -19.506000518798828 y:11.520999908447266 z:27.60099983215332
- atom: CG x: -18.007999420166016 y:11.626999855041504 z:27.636999130249023
- atom: CD1 x: -17.367000579833984 y:12.77299976348877 z:27.167999267578125
- atom: CD2 x: -17.236000061035156 y:10.593999862670898 z:28.156999588012695
- atom: CE1 x: -15.989999771118164 y:12.890000343322754 z:27.218000411987305
- atom: CE2 x: -15.854999542236328 y:10.704000473022461 z:28.211000442504883
- atom: CZ x: -15.229999542236328 y:11.857000350952148 z:27.739999771118164
- residue THR has 7 atoms
- atom: N x: -18.934999465942383 y:12.182999610900879 z:24.211000442504883
- atom: CA x: -18.201000213623047 y:11.710000038146973 z:23.042999267578125
- atom: C x: -17.07200050354004 y:12.701000213623047 z:22.77400016784668
- atom: O x: -16.788000106811523 y:13.565999984741211 z:23.59600067138672
- atom: CB x: -19.110000610351562 y:11.645999908447266 z:21.799999237060547
- atom: OG1 x: -19.204999923706055 y:12.947999954223633 z:21.209999084472656
- atom: CG2 x: -20.5049991607666 y:11.180000305175781 z:22.18400001525879
- residue SER has 6 atoms
- atom: N x: -16.42799949645996 y:12.579000473022461 z:21.625999450683594
- atom: CA x: -15.345999717712402 y:13.48900032043457 z:21.288000106811523
- atom: C x: -15.72700023651123 y:14.932999610900879 z:21.63599967956543
- atom: O x: -15.057999610900879 y:15.576000213623047 z:22.45199966430664
- atom: CB x: -15.020000457763672 y:13.368000030517578 z:19.798999786376953
- atom: OG x: -14.71500015258789 y:12.024999618530273 z:19.464000701904297
- residue GLU has 9 atoms
- atom: N x: -16.80699920654297 y:15.42199993133545 z:21.023000717163086
- atom: CA x: -17.32900047302246 y:16.783000946044922 z:21.238000869750977
- atom: C x: -17.645000457763672 y:17.051000595092773 z:22.7189998626709
- atom: O x: -16.93400001525879 y:17.798999786376953 z:23.395999908447266
- atom: CB x: -18.590999603271484 y:16.97100067138672 z:20.389999389648438
- atom: CG x: -19.5049991607666 y:15.744000434875488 z:20.41900062561035
- atom: CD x: -20.75200080871582 y:15.880999565124512 z:19.555999755859375
- atom: OE1 x: -20.6200008392334 y:16.08300018310547 z:18.32900047302246
- atom: OE2 x: -21.8700008392334 y:15.776000022888184 z:20.10700035095215
- residue SER has 6 atoms
- atom: N x: -18.738000869750977 y:16.454999923706055 z:23.19099998474121
- atom: CA x: -19.179000854492188 y:16.530000686645508 z:24.583999633789062
- atom: C x: -19.48200035095215 y:17.86400032043457 z:25.290000915527344
- atom: O x: -20.475000381469727 y:17.961999893188477 z:26.020999908447266
- atom: CB x: -18.18199920654297 y:15.746000289916992 z:25.451000213623047
- atom: OG x: -16.867000579833984 y:16.288999557495117 z:25.367000579833984
- residue THR has 7 atoms
- atom: N x: -18.636999130249023 y:18.87299919128418 z:25.0939998626709
- atom: CA x: -18.801000595092773 y:20.159000396728516 z:25.781999588012695
- atom: C x: -18.413999557495117 y:19.841999053955078 z:27.225000381469727
- atom: O x: -18.558000564575195 y:20.67099952697754 z:28.12700080871582
- atom: CB x: -20.27400016784668 y:20.67799949645996 z:25.78700065612793
- atom: OG1 x: -20.80299949645996 y:20.674999237060547 z:24.45400047302246
- atom: CG2 x: -20.33799934387207 y:22.104999542236328 z:26.34600067138672
- residue LYS has 9 atoms
- atom: N x: -17.930999755859375 y:18.615999221801758 z:27.42099952697754
- atom: CA x: -17.509000778198242 y:18.101999282836914 z:28.725000381469727
- atom: C x: -18.625 y:18.062000274658203 z:29.767000198364258
- atom: O x: -19.027000427246094 y:19.090999603271484 z:30.31800079345703
- atom: CB x: -16.312000274658203 y:18.898000717163086 z:29.25200080871582
- atom: CG x: -14.979999542236328 y:18.506999969482422 z:28.608999252319336
- atom: CD x: -14.986000061035156 y:18.663999557495117 z:27.079999923706055
- atom: CE x: -13.618000030517578 y:18.32900047302246 z:26.48699951171875
- atom: NZ x: -13.541999816894531 y:18.54800033569336 z:25.016000747680664
- residue GLN has 9 atoms
- atom: N x: -19.118999481201172 y:16.850000381469727 z:30.014999389648438
- atom: CA x: -20.18400001525879 y:16.606000900268555 z:30.979000091552734
- atom: C x: -19.53700065612793 y:16.427000045776367 z:32.35100173950195
- atom: O x: -18.87299919128418 y:15.42199993133545 z:32.61600112915039
- atom: CB x: -20.966999053955078 y:15.35099983215332 z:30.586000442504883
- atom: CG x: -22.29400062561035 y:15.206999778747559 z:31.299999237060547
- atom: CD x: -23.20400047302246 y:16.392000198364258 z:31.062000274658203
- atom: OE1 x: -23.5310001373291 y:16.709999084472656 z:29.92300033569336
- atom: NE2 x: -23.618000030517578 y:17.05500030517578 z:32.137001037597656
- residue PRO has 7 atoms
- atom: N x: -19.719999313354492 y:17.417999267578125 z:33.23699951171875
- atom: CA x: -19.187000274658203 y:17.464000701904297 z:34.60200119018555
- atom: C x: -19.802000045776367 y:16.465999603271484 z:35.58100128173828
- atom: O x: -20.979000091552734 y:16.569000244140625 z:35.93000030517578
- atom: CB x: -19.45400047302246 y:18.90399932861328 z:35.012001037597656
- atom: CG x: -20.766000747680664 y:19.173999786376953 z:34.35300064086914
- atom: CD x: -20.55299949645996 y:18.604999542236328 z:32.970001220703125
- residue LYS has 9 atoms
- atom: N x: -18.98699951171875 y:15.51099967956543 z:36.02000045776367
- atom: CA x: -19.39900016784668 y:14.482000350952148 z:36.97200012207031
- atom: C x: -20.075000762939453 y:13.255000114440918 z:36.362998962402344
- atom: O x: -20.964000701904297 y:12.654999732971191 z:36.9739990234375
- atom: CB x: -20.297000885009766 y:15.085000038146973 z:38.06100082397461
- atom: CG x: -19.63599967956543 y:16.225000381469727 z:38.82099914550781
- atom: CD x: -20.55299949645996 y:16.827999114990234 z:39.87300109863281
- atom: CE x: -19.891000747680664 y:18.023000717163086 z:40.55099868774414
- atom: NZ x: -20.739999771118164 y:18.586000442504883 z:41.63999938964844
- residue LEU has 8 atoms
- atom: N x: -19.663999557495117 y:12.876999855041504 z:35.15700149536133
- atom: CA x: -20.231000900268555 y:11.685999870300293 z:34.54800033569336
- atom: C x: -19.660999298095703 y:10.541000366210938 z:35.36899948120117
- atom: O x: -20.357999801635742 y:9.57800006866455 z:35.69499969482422
- atom: CB x: -19.77899932861328 y:11.529000282287598 z:33.09600067138672
- atom: CG x: -20.839000701904297 y:11.605999946594238 z:31.996999740600586
- atom: CD1 x: -20.340999603271484 y:10.803999900817871 z:30.812000274658203
- atom: CD2 x: -22.187000274658203 y:11.057000160217285 z:32.479000091552734
- residue LEU has 8 atoms
- atom: N x: -18.3799991607666 y:10.666000366210938 z:35.709999084472656
- atom: CA x: -17.700000762939453 y:9.656000137329102 z:36.494998931884766
- atom: C x: -17.95599937438965 y:9.869999885559082 z:37.96799850463867
- atom: O x: -18.058000564575195 y:8.918999671936035 z:38.729000091552734
- atom: CB x: -16.20400047302246 y:9.694000244140625 z:36.222999572753906
- atom: CG x: -15.364999771118164 y:8.619000434875488 z:36.91600036621094
- atom: CD1 x: -15.930000305175781 y:7.230999946594238 z:36.617000579833984
- atom: CD2 x: -13.913000106811523 y:8.743000030517578 z:36.44200134277344
- residue HIS has 10 atoms
- atom: N x: -18.06999969482422 y:11.121999740600586 z:38.37699890136719
- atom: CA x: -18.312000274658203 y:11.401000022888184 z:39.779998779296875
- atom: C x: -19.5310001373291 y:10.654999732971191 z:40.31700134277344
- atom: O x: -19.44099998474121 y:9.95300006866455 z:41.316001892089844
- atom: CB x: -18.496999740600586 y:12.901000022888184 z:40.000999450683594
- atom: CG x: -18.812000274658203 y:13.258999824523926 z:41.41999816894531
- atom: ND1 x: -17.89900016784668 y:13.11299991607666 z:42.44200134277344
- atom: CD2 x: -19.959999084472656 y:13.682999610900879 z:41.99700164794922
- atom: CE1 x: -18.474000930786133 y:13.425000190734863 z:43.59000015258789
- atom: NE2 x: -19.725000381469727 y:13.77400016784668 z:43.347999572753906
- residue GLN has 9 atoms
- atom: N x: -20.66900062561035 y:10.812999725341797 z:39.64699935913086
- atom: CA x: -21.92300033569336 y:10.175999641418457 z:40.055999755859375
- atom: C x: -21.83099937438965 y:8.678000450134277 z:40.29800033569336
- atom: O x: -22.37700080871582 y:8.17300033569336 z:41.268001556396484
- atom: CB x: -23.016000747680664 y:10.432000160217285 z:39.018001556396484
- atom: CG x: -23.368000030517578 y:11.890999794006348 z:38.82099914550781
- atom: CD x: -24.46299934387207 y:12.07699966430664 z:37.78099822998047
- atom: OE1 x: -25.597999572753906 y:11.62600040435791 z:37.9640007019043
- atom: NE2 x: -24.124000549316406 y:12.73799991607666 z:36.67900085449219
- residue VAL has 7 atoms
- atom: N x: -21.155000686645508 y:7.968999862670898 z:39.409000396728516
- atom: CA x: -21.006000518798828 y:6.53000020980835 z:39.54600143432617
- atom: C x: -20.003000259399414 y:6.197000026702881 z:40.6510009765625
- atom: O x: -20.218000411987305 y:5.28000020980835 z:41.4379997253418
- atom: CB x: -20.562999725341797 y:5.888000011444092 z:38.20899963378906
- atom: CG1 x: -20.05699920654297 y:4.473999977111816 z:38.441001892089844
- atom: CG2 x: -21.73699951171875 y:5.868000030517578 z:37.24599838256836
- residue VAL has 7 atoms
- atom: N x: -18.913999557495117 y:6.947000026702881 z:40.720001220703125
- atom: CA x: -17.933000564575195 y:6.690000057220459 z:41.75199890136719
- atom: C x: -18.690000534057617 y:6.857999801635742 z:43.042999267578125
- atom: O x: -18.660999298095703 y:6.008999824523926 z:43.92399978637695
- atom: CB x: -16.781999588012695 y:7.709000110626221 z:41.72200012207031
- atom: CG1 x: -15.850000381469727 y:7.456999778747559 z:42.880001068115234
- atom: CG2 x: -16.020000457763672 y:7.6020002365112305 z:40.41999816894531
- residue GLU has 9 atoms
- atom: N x: -19.398000717163086 y:7.968999862670898 z:43.132999420166016
- atom: CA x: -20.167999267578125 y:8.276000022888184 z:44.319000244140625
- atom: C x: -21.114999771118164 y:7.140999794006348 z:44.65999984741211
- atom: O x: -21.065000534057617 y:6.604000091552734 z:45.75699996948242
- atom: CB x: -20.961000442504883 y:9.5600004196167 z:44.09700012207031
- atom: CG x: -21.607999801635742 y:10.109999656677246 z:45.33399963378906
- atom: CD x: -22.40399932861328 y:11.36299991607666 z:45.042999267578125
- atom: OE1 x: -23.4689998626709 y:11.255999565124512 z:44.395999908447266
- atom: OE2 x: -21.961999893188477 y:12.458000183105469 z:45.452999114990234
- residue GLN has 9 atoms
- atom: N x: -21.9689998626709 y:6.763000011444092 z:43.715999603271484
- atom: CA x: -22.940000534057617 y:5.702000141143799 z:43.96500015258789
- atom: C x: -22.292999267578125 y:4.389999866485596 z:44.44900131225586
- atom: O x: -22.858999252319336 y:3.684000015258789 z:45.28799819946289
- atom: CB x: -23.791000366210938 y:5.4720001220703125 z:42.70100021362305
- atom: CG x: -24.867000579833984 y:4.372000217437744 z:42.79600143432617
- atom: CD x: -26.01300048828125 y:4.7129998207092285 z:43.7400016784668
- atom: OE1 x: -25.9689998626709 y:5.706999778747559 z:44.46099853515625
- atom: NE2 x: -27.04400062561035 y:3.874000072479248 z:43.742000579833984
- residue LEU has 8 atoms
- atom: N x: -21.11199951171875 y:4.065000057220459 z:43.93299865722656
- atom: CA x: -20.437000274658203 y:2.8420000076293945 z:44.3489990234375
- atom: C x: -19.951000213623047 y:2.9660000801086426 z:45.7859992980957
- atom: O x: -19.659000396728516 y:1.9639999866485596 z:46.42499923706055
- atom: CB x: -19.267000198364258 y:2.5199999809265137 z:43.4109992980957
- atom: CG x: -19.673999786376953 y:1.9570000171661377 z:42.04199981689453
- atom: CD1 x: -18.472999572753906 y:1.777999997138977 z:41.165000915527344
- atom: CD2 x: -20.384000778198242 y:0.628000020980835 z:42.23400115966797
- residue GLN has 9 atoms
- atom: N x: -19.868000030517578 y:4.198999881744385 z:46.28799819946289
- atom: CA x: -19.44499969482422 y:4.461999893188477 z:47.66899871826172
- atom: C x: -20.687999725341797 y:4.511000156402588 z:48.558998107910156
- atom: O x: -20.60099983215332 y:4.388999938964844 z:49.7869987487793
- atom: CB x: -18.71500015258789 y:5.809000015258789 z:47.7859992980957
- atom: CG x: -17.40399932861328 y:5.919000148773193 z:47.03499984741211
- atom: CD x: -16.65999984741211 y:7.2210001945495605 z:47.33300018310547
- atom: OE1 x: -17.259000778198242 y:8.29800033569336 z:47.375999450683594
- atom: NE2 x: -15.347000122070312 y:7.123000144958496 z:47.527000427246094
- residue LYS has 9 atoms
- atom: N x: -21.84000015258789 y:4.710000038146973 z:47.92300033569336
- atom: CA x: -23.118000030517578 y:4.790999889373779 z:48.62099838256836
- atom: C x: -23.485000610351562 y:3.427999973297119 z:49.20899963378906
- atom: O x: -23.861000061035156 y:3.321000099182129 z:50.37699890136719
- atom: CB x: -24.209999084472656 y:5.244999885559082 z:47.6510009765625
- atom: CG x: -25.493000030517578 y:5.758999824523926 z:48.308998107910156
- atom: CD x: -25.284000396728516 y:7.151000022888184 z:48.91600036621094
- atom: CE x: -26.61400032043457 y:7.869999885559082 z:49.20199966430664
- atom: NZ x: -27.492000579833984 y:7.140999794006348 z:50.17300033569336
- residue VAL has 7 atoms
- atom: N x: -23.381000518798828 y:2.38700008392334 z:48.391998291015625
- atom: CA x: -23.690000534057617 y:1.0379999876022339 z:48.84199905395508
- atom: C x: -22.56100082397461 y:0.5149999856948853 z:49.72800064086914
- atom: O x: -22.729000091552734 y:-0.4690000116825104 z:50.45100021362305
- atom: CB x: -23.85300064086914 y:0.07800000160932541 z:47.645999908447266
- atom: CG1 x: -25.08300018310547 y:0.44999998807907104 z:46.845001220703125
- atom: CG2 x: -22.618999481201172 y:0.13099999725818634 z:46.76300048828125
- residue HIS has 10 atoms
- atom: N x: -21.413999557495117 y:1.187999963760376 z:49.66400146484375
- atom: CA x: -20.229000091552734 y:0.8109999895095825 z:50.428001403808594
- atom: C x: -19.409000396728516 y:-0.27000001072883606 z:49.709999084472656
- atom: O x: -18.84000015258789 y:-1.152999997138977 z:50.35200119018555
- atom: CB x: -20.610000610351562 y:0.33399999141693115 z:51.8390007019043
- atom: CG x: -21.040000915527344 y:1.437000036239624 z:52.7599983215332
- atom: ND1 x: -22.143999099731445 y:1.3350000381469727 z:53.582000732421875
- atom: CD2 x: -20.5 y:2.6549999713897705 z:53.0099983215332
- atom: CE1 x: -22.266000747680664 y:2.440000057220459 z:54.297000885009766
- atom: NE2 x: -21.281999588012695 y:3.257999897003174 z:53.96900177001953
- residue PHE has 11 atoms
- atom: N x: -19.360000610351562 y:-0.20000000298023224 z:48.37799835205078
- atom: CA x: -18.570999145507812 y:-1.1480000019073486 z:47.59000015258789
- atom: C x: -17.174999237060547 y:-0.5789999961853027 z:47.39500045776367
- atom: O x: -16.20800018310547 y:-1.3270000219345093 z:47.3650016784668
- atom: CB x: -19.187999725341797 y:-1.3940000534057617 z:46.220001220703125
- atom: CG x: -18.344999313354492 y:-2.2720000743865967 z:45.32500076293945
- atom: CD1 x: -18.054000854492188 y:-3.5820000171661377 z:45.685001373291016
- atom: CD2 x: -17.84600067138672 y:-1.784999966621399 z:44.11899948120117
- atom: CE1 x: -17.288999557495117 y:-4.38700008392334 z:44.86800003051758
- atom: CE2 x: -17.076000213623047 y:-2.5869998931884766 z:43.29399871826172
- atom: CZ x: -16.79800033569336 y:-3.8910000324249268 z:43.67100143432617
- residue ILE has 8 atoms
- atom: N x: -17.072999954223633 y:0.7419999837875366 z:47.24300003051758
- atom: CA x: -15.77400016784668 y:1.3860000371932983 z:47.08300018310547
- atom: C x: -15.430999755859375 y:1.9910000562667847 z:48.42300033569336
- atom: O x: -16.05900001525879 y:2.953000068664551 z:48.85200119018555
- atom: CB x: -15.774999618530273 y:2.503000020980835 z:45.97999954223633
- atom: CG1 x: -15.883000373840332 y:1.871000051498413 z:44.58399963378906
- atom: CG2 x: -14.479000091552734 y:3.2899999618530273 z:46.02199935913086
- atom: CD1 x: -15.685999870300293 y:2.8510000705718994 z:43.44300079345703
- residue THR has 7 atoms
- atom: N x: -14.427000045776367 y:1.4140000343322754 z:49.073001861572266
- atom: CA x: -13.984999656677246 y:1.8559999465942383 z:50.388999938964844
- atom: C x: -12.88599967956543 y:2.9140000343322754 z:50.402000427246094
- atom: O x: -12.743000030517578 y:3.63100004196167 z:51.39099884033203
- atom: CB x: -13.479000091552734 y:0.6610000133514404 z:51.25299835205078
- atom: OG1 x: -12.404000282287598 y:-0.0020000000949949026 z:50.58300018310547
- atom: CG2 x: -14.581000328063965 y:-0.32600000500679016 z:51.51499938964844
- residue ASP has 8 atoms
- atom: N x: -12.095000267028809 y:3.01200008392334 z:49.3390007019043
- atom: CA x: -11.015000343322754 y:4.000999927520752 z:49.31800079345703
- atom: C x: -10.654000282287598 y:4.551000118255615 z:47.93299865722656
- atom: O x: -10.78600025177002 y:3.868000030517578 z:46.915000915527344
- atom: CB x: -9.744000434875488 y:3.4130001068115234 z:49.9630012512207
- atom: CG x: -10.015000343322754 y:2.7699999809265137 z:51.31800079345703
- atom: OD1 x: -10.675000190734863 y:1.7079999446868896 z:51.349998474121094
- atom: OD2 x: -9.574000358581543 y:3.3239998817443848 z:52.35100173950195
- residue THR has 7 atoms
- atom: N x: -10.157999992370605 y:5.7870001792907715 z:47.93199920654297
- atom: CA x: -9.746999740600586 y:6.508999824523926 z:46.73899841308594
- atom: C x: -8.234999656677246 y:6.723999977111816 z:46.68000030517578
- atom: O x: -7.664999961853027 y:7.307000160217285 z:47.59000015258789
- atom: CB x: -10.407999992370605 y:7.89300012588501 z:46.715999603271484
- atom: OG1 x: -11.79800033569336 y:7.748000144958496 z:46.40999984741211
- atom: CG2 x: -9.73799991607666 y:8.803000450134277 z:45.6879997253418
- residue LEU has 8 atoms
- atom: N x: -7.583000183105469 y:6.265999794006348 z:45.612998962402344
- atom: CA x: -6.146999835968018 y:6.489999771118164 z:45.47800064086914
- atom: C x: -5.991000175476074 y:7.702000141143799 z:44.57099914550781
- atom: O x: -5.010000228881836 y:8.442000389099121 z:44.667999267578125
- atom: CB x: -5.427999973297119 y:5.298999786376953 z:44.83100128173828
- atom: CG x: -5.5980000495910645 y:3.871999979019165 z:45.35599899291992
- atom: CD1 x: -4.579999923706055 y:2.9730000495910645 z:44.696998596191406
- atom: CD2 x: -5.414999961853027 y:3.8259999752044678 z:46.847999572753906
- residue SER has 6 atoms
- atom: N x: -6.9710001945495605 y:7.901000022888184 z:43.694000244140625
- atom: CA x: -6.947999954223633 y:9.012999534606934 z:42.74399948120117
- atom: C x: -8.258999824523926 y:9.112000465393066 z:41.98500061035156
- atom: O x: -8.848999977111816 y:8.095000267028809 z:41.61399841308594
- atom: CB x: -5.817999839782715 y:8.831999778747559 z:41.74100112915039
- atom: OG x: -6.002999782562256 y:7.638999938964844 z:41.01300048828125
- residue LYS has 9 atoms
- atom: N x: -8.70300006866455 y:10.340999603271484 z:41.74800109863281
- atom: CA x: -9.954000473022461 y:10.569999694824219 z:41.04100036621094
- atom: C x: -9.918000221252441 y:11.857999801635742 z:40.21900177001953
- atom: O x: -9.762999534606934 y:12.961999893188477 z:40.74599838256836
- atom: CB x: -11.107999801635742 y:10.605999946594238 z:42.04499816894531
- atom: CG x: -12.496000289916992 y:10.840999603271484 z:41.47100067138672
- atom: CD x: -13.52299976348877 y:10.961000442504883 z:42.61000061035156
- atom: CE x: -14.862000465393066 y:11.560999870300293 z:42.15599822998047
- atom: NZ x: -14.739999771118164 y:12.946999549865723 z:41.57699966430664
- residue GLY has 4 atoms
- atom: N x: -10.03600025177002 y:11.694000244140625 z:38.90800094604492
- atom: CA x: -10.041999816894531 y:12.824000358581543 z:38.00600051879883
- atom: C x: -11.36400032043457 y:12.800000190734863 z:37.26900100708008
- atom: O x: -12.270999908447266 y:12.064000129699707 z:37.66899871826172
- residue GLU has 9 atoms
- atom: N x: -11.475000381469727 y:13.581999778747559 z:36.19599914550781
- atom: CA x: -12.708999633789062 y:13.637999534606934 z:35.41299819946289
- atom: C x: -12.991000175476074 y:12.404999732971191 z:34.569000244140625
- atom: O x: -14.145000457763672 y:12.163000106811523 z:34.23899841308594
- atom: CB x: -12.729000091552734 y:14.871000289916992 z:34.49399948120117
- atom: CG x: -12.994000434875488 y:16.202999114990234 z:35.20000076293945
- atom: CD x: -14.388999938964844 y:16.284000396728516 z:35.803001403808594
- atom: OE1 x: -14.821999549865723 y:15.300000190734863 z:36.452999114990234
- atom: OE2 x: -15.04800033569336 y:17.33300018310547 z:35.64099884033203
- residue THR has 7 atoms
- atom: N x: -11.963000297546387 y:11.63700008392334 z:34.209999084472656
- atom: CA x: -12.175000190734863 y:10.432999610900879 z:33.39699935913086
- atom: C x: -11.51200008392334 y:9.157999992370605 z:33.95399856567383
- atom: O x: -11.673999786376953 y:8.072999954223633 z:33.38600158691406
- atom: CB x: -11.6850004196167 y:10.630000114440918 z:31.908000946044922
- atom: OG1 x: -10.255999565124512 y:10.644000053405762 z:31.856000900268555
- atom: CG2 x: -12.185999870300293 y:11.944999694824219 z:31.336999893188477
- residue LYS has 9 atoms
- atom: N x: -10.781000137329102 y:9.277000427246094 z:35.06399917602539
- atom: CA x: -10.095999717712402 y:8.119999885559082 z:35.63800048828125
- atom: C x: -10.126999855041504 y:8.034000396728516 z:37.154998779296875
- atom: O x: -9.835000038146973 y:9.008000373840332 z:37.849998474121094
- atom: CB x: -8.642999649047852 y:8.10099983215332 z:35.18199920654297
- atom: CG x: -7.811999797821045 y:7.0980000495910645 z:35.92300033569336
- atom: CD x: -6.353000164031982 y:7.2220001220703125 z:35.58100128173828
- atom: CE x: -5.540999889373779 y:6.206999778747559 z:36.380001068115234
- atom: NZ x: -4.105000019073486 y:6.232999801635742 z:36.0
- residue PHE has 11 atoms
- atom: N x: -10.454000473022461 y:6.8460001945495605 z:37.65800094604492
- atom: CA x: -10.522000312805176 y:6.5879998207092285 z:39.090999603271484
- atom: C x: -9.812000274658203 y:5.298999786376953 z:39.49700164794922
- atom: O x: -10.163999557495117 y:4.2129998207092285 z:39.051998138427734
- atom: CB x: -11.984000205993652 y:6.501999855041504 z:39.547000885009766
- atom: CG x: -12.156000137329102 y:6.059999942779541 z:40.986000061035156
- atom: CD1 x: -11.930999755859375 y:6.936999797821045 z:42.0359992980957
- atom: CD2 x: -12.531000137329102 y:4.756999969482422 z:41.2859992980957
- atom: CE1 x: -12.079000473022461 y:6.519000053405762 z:43.36600112915039
- atom: CE2 x: -12.678999900817871 y:4.329999923706055 z:42.61800003051758
- atom: CZ x: -12.45300006866455 y:5.210999965667725 z:43.652000427246094
- residue MET has 8 atoms
- atom: N x: -8.805999755859375 y:5.428999900817871 z:40.35100173950195
- atom: CA x: -8.081999778747559 y:4.275000095367432 z:40.875
- atom: C x: -8.491999626159668 y:4.190000057220459 z:42.34199905395508
- atom: O x: -8.345999717712402 y:5.159999847412109 z:43.09000015258789
- atom: CB x: -6.578999996185303 y:4.486000061035156 z:40.77000045776367
- atom: CG x: -6.034999847412109 y:4.218999862670898 z:39.39500045776367
- atom: SD x: -4.295000076293945 y:4.629000186920166 z:39.29399871826172
- atom: CE x: -3.693000078201294 y:3.8350000381469727 z:40.74300003051758
- residue GLY has 4 atoms
- atom: N x: -9.01200008392334 y:3.0390000343322754 z:42.7599983215332
- atom: CA x: -9.442999839782715 y:2.933000087738037 z:44.13600158691406
- atom: C x: -9.482999801635742 y:1.5640000104904175 z:44.742000579833984
- atom: O x: -8.850000381469727 y:0.6309999823570251 z:44.27000045776367
- residue VAL has 7 atoms
- atom: N x: -10.26200008392334 y:1.4559999704360962 z:45.80400085449219
- atom: CA x: -10.385000228881836 y:0.21899999678134918 z:46.54199981689453
- atom: C x: -11.847000122070312 y:-0.14499999582767487 z:46.7239990234375
- atom: O x: -12.666000366210938 y:0.7099999785423279 z:47.07899856567383
- atom: CB x: -9.720000267028809 y:0.36800000071525574 z:47.94599914550781
- atom: CG1 x: -9.960000038146973 y:-0.8809999823570251 z:48.790000915527344
- atom: CG2 x: -8.218000411987305 y:0.6430000066757202 z:47.7869987487793
- residue CYS has 6 atoms
- atom: N x: -12.168999671936035 y:-1.4140000343322754 z:46.46799850463867
- atom: CA x: -13.529000282287598 y:-1.909000039100647 z:46.63399887084961
- atom: C x: -13.513999938964844 y:-3.187000036239624 z:47.46699905395508
- atom: O x: -12.456000328063965 y:-3.759000062942505 z:47.72200012207031
- atom: CB x: -14.20300006866455 y:-2.1549999713897705 z:45.277000427246094
- atom: SG x: -13.541000366210938 y:-3.48799991607666 z:44.275001525878906
- residue GLN has 9 atoms
- atom: N x: -14.690999984741211 y:-3.625999927520752 z:47.900001525878906
- atom: CA x: -14.809000015258789 y:-4.818999767303467 z:48.72600173950195
- atom: C x: -16.209999084472656 y:-5.434999942779541 z:48.64799880981445
- atom: O x: -17.215999603271484 y:-4.756999969482422 z:48.869998931884766
- atom: CB x: -14.467000007629395 y:-4.459000110626221 z:50.178001403808594
- atom: CG x: -14.763999938964844 y:-5.531000137329102 z:51.202999114990234
- atom: CD x: -14.333999633789062 y:-5.130000114440918 z:52.604000091552734
- atom: OE1 x: -14.741000175476074 y:-4.0879998207092285 z:53.11800003051758
- atom: NE2 x: -13.508000373840332 y:-5.961999893188477 z:53.231998443603516
- residue LEU has 8 atoms
- atom: N x: -16.267000198364258 y:-6.71999979019165 z:48.31399917602539
- atom: CA x: -17.533000946044922 y:-7.429999828338623 z:48.22800064086914
- atom: C x: -18.14900016784668 y:-7.479000091552734 z:49.61899948120117
- atom: O x: -17.454999923706055 y:-7.729000091552734 z:50.595001220703125
- atom: CB x: -17.297000885009766 y:-8.852999687194824 z:47.722999572753906
- atom: CG x: -16.979000091552734 y:-9.00100040435791 z:46.236000061035156
- atom: CD1 x: -16.43600082397461 y:-10.39799976348877 z:45.946998596191406
- atom: CD2 x: -18.249000549316406 y:-8.720000267028809 z:45.43000030517578
- residue PRO has 7 atoms
- atom: N x: -19.459999084472656 y:-7.241000175476074 z:49.733001708984375
- atom: CA x: -20.0939998626709 y:-7.28000020980835 z:51.053001403808594
- atom: C x: -20.091999053955078 y:-8.692999839782715 z:51.62099838256836
- atom: O x: -20.04400062561035 y:-9.668000221252441 z:50.87300109863281
- atom: CB x: -21.49799919128418 y:-6.7829999923706055 z:50.763999938964844
- atom: CG x: -21.75200080871582 y:-7.359000205993652 z:49.42300033569336
- atom: CD x: -20.466999053955078 y:-7.035999774932861 z:48.683998107910156
- residue SER has 6 atoms
- atom: N x: -20.14299964904785 y:-8.800000190734863 z:52.944000244140625
- atom: CA x: -20.150999069213867 y:-10.104999542236328 z:53.59000015258789
- atom: C x: -21.516000747680664 y:-10.470999717712402 z:54.14400100708008
- atom: O x: -22.22800064086914 y:-9.626999855041504 z:54.69300079345703
- atom: CB x: -19.11400032043457 y:-10.157999992370605 z:54.71099853515625
- atom: OG x: -17.80299949645996 y:-10.243000030517578 z:54.176998138427734
- residue LYS has 9 atoms
- atom: N x: -21.854999542236328 y:-11.748000144958496 z:53.99100112915039
- atom: CA x: -23.124000549316406 y:-12.317000389099121 z:54.44300079345703
- atom: C x: -23.631999969482422 y:-11.708000183105469 z:55.74700164794922
- atom: O x: -24.525999069213867 y:-10.859999656677246 z:55.74100112915039
- atom: CB x: -22.985000610351562 y:-13.836000442504883 z:54.61800003051758
- atom: CG x: -22.371000289916992 y:-14.569000244140625 z:53.42399978637695
- atom: CD x: -20.875999450683594 y:-14.298999786376953 z:53.28200149536133
- atom: CE x: -20.301000595092773 y:-14.970999717712402 z:52.04600143432617
- atom: NZ x: -18.827999114990234 y:-14.781999588012695 z:51.97600173950195
- residue ASN has 8 atoms
- atom: N x: -23.066999435424805 y:-12.161999702453613 z:56.861000061035156
- atom: CA x: -23.440000534057617 y:-11.675999641418457 z:58.18600082397461
- atom: C x: -22.19700050354004 y:-11.550000190734863 z:59.04600143432617
- atom: O x: -21.503999710083008 y:-10.529999732971191 z:59.018001556396484
- atom: CB x: -24.42300033569336 y:-12.637999534606934 z:58.86199951171875
- atom: CG x: -25.77899932861328 y:-12.663999557495117 z:58.180999755859375
- atom: OD1 x: -25.885000228881836 y:-12.98799991607666 z:56.99300003051758
- atom: ND2 x: -26.826000213623047 y:-12.321999549865723 z:58.93000030517578
- residue ASP has 8 atoms
- atom: N x: -21.91200065612793 y:-12.593999862670898 z:59.8120002746582
- atom: CA x: -20.743000030517578 y:-12.5649995803833 z:60.667999267578125
- atom: C x: -19.663999557495117 y:-13.559000015258789 z:60.22200012207031
- atom: O x: -19.101999282836914 y:-14.295999526977539 z:61.034000396728516
- atom: CB x: -21.1560001373291 y:-12.817000389099121 z:62.12099838256836
- atom: CG x: -20.41699981689453 y:-11.913000106811523 z:63.0989990234375
- atom: OD1 x: -20.413000106811523 y:-10.680999755859375 z:62.880001068115234
- atom: OD2 x: -19.844999313354492 y:-12.430999755859375 z:64.08399963378906
- residue GLU has 9 atoms
- atom: N x: -19.38800048828125 y:-13.57699966430664 z:58.917999267578125
- atom: CA x: -18.354000091552734 y:-14.447999954223633 z:58.375
- atom: C x: -17.06800079345703 y:-13.638999938964844 z:58.441001892089844
- atom: O x: -16.729000091552734 y:-13.08899974822998 z:59.4900016784668
- atom: CB x: -18.63800048828125 y:-14.824999809265137 z:56.91699981689453
- atom: CG x: -19.93899917602539 y:-15.576000213623047 z:56.67300033569336
- atom: CD x: -21.152000427246094 y:-14.701000213623047 z:56.88600158691406
- atom: OE1 x: -21.05900001525879 y:-13.493000030517578 z:56.584999084472656
- atom: OE2 x: -22.195999145507812 y:-15.218000411987305 z:57.3380012512207
- residue LYS has 9 atoms
- atom: N x: -16.361000061035156 y:-13.54699993133545 z:57.319000244140625
- atom: CA x: -15.116000175476074 y:-12.793999671936035 z:57.284000396728516
- atom: C x: -15.039999961853027 y:-11.869000434875488 z:56.07699966430664
- atom: O x: -15.192000389099121 y:-12.310999870300293 z:54.941001892089844
- atom: CB x: -13.92300033569336 y:-13.751999855041504 z:57.27399826049805
- atom: CG x: -12.5600004196167 y:-13.060999870300293 z:57.27399826049805
- atom: CD x: -11.418999671936035 y:-14.069999694824219 z:57.36899948120117
- atom: CE x: -11.454000473022461 y:-15.064000129699707 z:56.21500015258789
- atom: NZ x: -10.368000030517578 y:-16.07900047302246 z:56.319000244140625
- residue GLU has 9 atoms
- atom: N x: -14.805000305175781 y:-10.585000038146973 z:56.340999603271484
- atom: CA x: -14.687000274658203 y:-9.57800006866455 z:55.292999267578125
- atom: C x: -13.902999877929688 y:-10.180000305175781 z:54.125999450683594
- atom: O x: -12.942000389099121 y:-10.918999671936035 z:54.34199905395508
- atom: CB x: -13.907999992370605 y:-8.350000381469727 z:55.79999923706055
- atom: CG x: -14.197999954223633 y:-7.88700008392334 z:57.22200012207031
- atom: CD x: -13.293000221252441 y:-6.72599983215332 z:57.652000427246094
- atom: OE1 x: -13.357000350952148 y:-5.650000095367432 z:57.01900100708008
- atom: OE2 x: -12.512999534606934 y:-6.883999824523926 z:58.62099838256836
- residue TYR has 12 atoms
- atom: N x: -14.315999984741211 y:-9.883000373840332 z:52.895999908447266
- atom: CA x: -13.588000297546387 y:-10.373000144958496 z:51.72700119018555
- atom: C x: -12.331000328063965 y:-9.508000373840332 z:51.678001403808594
- atom: O x: -12.291000366210938 y:-8.444000244140625 z:52.292999267578125
- atom: CB x: -14.402000427246094 y:-10.159000396728516 z:50.44300079345703
- atom: CG x: -15.472000122070312 y:-11.201000213623047 z:50.17599868774414
- atom: CD1 x: -15.234999656677246 y:-12.276000022888184 z:49.31800079345703
- atom: CD2 x: -16.729000091552734 y:-11.105999946594238 z:50.7760009765625
- atom: CE1 x: -16.225000381469727 y:-13.22700023651123 z:49.064998626708984
- atom: CE2 x: -17.722999572753906 y:-12.048999786376953 z:50.52899932861328
- atom: CZ x: -17.465999603271484 y:-13.100000381469727 z:49.67599868774414
- atom: OH x: -18.458999633789062 y:-14.013999938964844 z:49.433998107910156
- residue PRO has 7 atoms
- atom: N x: -11.282999992370605 y:-9.958999633789062 z:50.972999572753906
- atom: CA x: -10.079999923706055 y:-9.125 z:50.917999267578125
- atom: C x: -10.451000213623047 y:-7.831999778747559 z:50.20899963378906
- atom: O x: -11.345000267028809 y:-7.835000038146973 z:49.35900115966797
- atom: CB x: -9.114999771118164 y:-9.960000038146973 z:50.07699966430664
- atom: CG x: -9.54699993133545 y:-11.354000091552734 z:50.3380012512207
- atom: CD x: -11.050999641418457 y:-11.258999824523926 z:50.32500076293945
- residue HIS has 10 atoms
- atom: N x: -9.805000305175781 y:-6.724999904632568 z:50.560001373291016
- atom: CA x: -10.095000267028809 y:-5.4710001945495605 z:49.867000579833984
- atom: C x: -9.428000450134277 y:-5.605000019073486 z:48.49399948120117
- atom: O x: -8.277999877929688 y:-6.047999858856201 z:48.395999908447266
- atom: CB x: -9.512999534606934 y:-4.267000198364258 z:50.619998931884766
- atom: CG x: -10.348999977111816 y:-3.796999931335449 z:51.77000045776367
- atom: ND1 x: -10.534000396728516 y:-4.544000148773193 z:52.91400146484375
- atom: CD2 x: -11.008000373840332 y:-2.631999969482422 z:51.97100067138672
- atom: CE1 x: -11.263999938964844 y:-3.8559999465942383 z:53.77299880981445
- atom: NE2 x: -11.564000129699707 y:-2.691999912261963 z:53.224998474121094
- residue ARG has 11 atoms
- atom: N x: -10.140999794006348 y:-5.234000205993652 z:47.4370002746582
- atom: CA x: -9.592000007629395 y:-5.364999771118164 z:46.08599853515625
- atom: C x: -9.380000114440918 y:-4.051000118255615 z:45.33399963378906
- atom: O x: -10.031000137329102 y:-3.0390000343322754 z:45.611000061035156
- atom: CB x: -10.51200008392334 y:-6.256999969482422 z:45.25899887084961
- atom: CG x: -10.968999862670898 y:-7.511000156402588 z:45.96200180053711
- atom: CD x: -9.836999893188477 y:-8.48799991607666 z:46.11199951171875
- atom: NE x: -10.319000244140625 y:-9.800999641418457 z:46.51900100708008
- atom: CZ x: -9.5600004196167 y:-10.888999938964844 z:46.54499816894531
- atom: NH1 x: -8.28499984741211 y:-10.8100004196167 z:46.19200134277344
- atom: NH2 x: -10.07699966430664 y:-12.057000160217285 z:46.902000427246094
- residue ARG has 11 atoms
- atom: N x: -8.461999893188477 y:-4.076000213623047 z:44.375
- atom: CA x: -8.197999954223633 y:-2.8940000534057617 z:43.558998107910156
- atom: C x: -9.196999549865723 y:-2.7960000038146973 z:42.402000427246094
- atom: O x: -9.545999526977539 y:-3.7890000343322754 z:41.75600051879883
- atom: CB x: -6.775000095367432 y:-2.9260001182556152 z:42.994998931884766
- atom: CG x: -5.702000141143799 y:-2.631999969482422 z:44.012001037597656
- atom: CD x: -5.521999835968018 y:-1.1480000019073486 z:44.24100112915039
- atom: NE x: -4.9710001945495605 y:-0.4950000047683716 z:43.0629997253418
- atom: CZ x: -5.703999996185303 y:0.11400000005960464 z:42.13800048828125
- atom: NH1 x: -7.0279998779296875 y:0.16500000655651093 z:42.25899887084961
- atom: NH2 x: -5.113999843597412 y:0.652999997138977 z:41.07899856567383
- residue ILE has 8 atoms
- atom: N x: -9.649999618530273 y:-1.5729999542236328 z:42.15399932861328
- atom: CA x: -10.597000122070312 y:-1.2949999570846558 z:41.0880012512207
- atom: C x: -10.175000190734863 y:-0.020999999716877937 z:40.34199905395508
- atom: O x: -9.767999649047852 y:0.9660000205039978 z:40.95800018310547
- atom: CB x: -12.017999649047852 y:-1.1380000114440918 z:41.68000030517578
- atom: CG1 x: -13.008999824523926 y:-0.7609999775886536 z:40.59199905395508
- atom: CG2 x: -12.031000137329102 y:-0.08799999952316284 z:42.75
- atom: CD1 x: -14.376999855041504 y:-0.5450000166893005 z:41.137001037597656
- residue ASP has 8 atoms
- atom: N x: -10.21500015258789 y:-0.07400000095367432 z:39.01499938964844
- atom: CA x: -9.883000373840332 y:1.069000005722046 z:38.1619987487793
- atom: C x: -11.109000205993652 y:1.2790000438690186 z:37.2760009765625
- atom: O x: -11.706999778747559 y:0.3100000023841858 z:36.80699920654297
- atom: CB x: -8.668000221252441 y:0.7850000262260437 z:37.270999908447266
- atom: CG x: -7.4070000648498535 y:0.503000020980835 z:38.064998626708984
- atom: OD1 x: -7.235000133514404 y:1.093000054359436 z:39.1510009765625
- atom: OD2 x: -6.578999996185303 y:-0.30000001192092896 z:37.5890007019043
- residue ILE has 8 atoms
- atom: N x: -11.487000465393066 y:2.5350000858306885 z:37.05500030517578
- atom: CA x: -12.64799976348877 y:2.8480000495910645 z:36.23400115966797
- atom: C x: -12.291999816894531 y:4.013000011444092 z:35.319000244140625
- atom: O x: -11.878999710083008 y:5.072000026702881 z:35.79499816894531
- atom: CB x: -13.845999717712402 y:3.256999969482422 z:37.12300109863281
- atom: CG1 x: -14.107999801635742 y:2.1679999828338623 z:38.16699981689453
- atom: CG2 x: -15.086999893188477 y:3.505000114440918 z:36.266998291015625
- atom: CD1 x: -15.119000434875488 y:2.562000036239624 z:39.233001708984375
- residue ARG has 11 atoms
- atom: N x: -12.4399995803833 y:3.8239998817443848 z:34.01100158691406
- atom: CA x: -12.133000373840332 y:4.89300012588501 z:33.064998626708984
- atom: C x: -13.319999694824219 y:5.24399995803833 z:32.17399978637695
- atom: O x: -13.930999755859375 y:4.36899995803833 z:31.548999786376953
- atom: CB x: -10.928000450134277 y:4.513000011444092 z:32.202999114990234
- atom: CG x: -9.746000289916992 y:4.109000205993652 z:33.055999755859375
- atom: CD x: -8.42300033569336 y:4.166999816894531 z:32.321998596191406
- atom: NE x: -7.381999969482422 y:3.446000099182129 z:33.055999755859375
- atom: CZ x: -6.118000030517578 y:3.3529999256134033 z:32.65599822998047
- atom: NH1 x: -5.738999843597412 y:3.947000026702881 z:31.527999877929688
- atom: NH2 x: -5.239999771118164 y:2.6530001163482666 z:33.36800003051758
- residue LEU has 8 atoms
- atom: N x: -13.649999618530273 y:6.5329999923706055 z:32.150001525878906
- atom: CA x: -14.743000030517578 y:7.039999961853027 z:31.347000122070312
- atom: C x: -14.180999755859375 y:7.448999881744385 z:29.990999221801758
- atom: O x: -13.451000213623047 y:8.435999870300293 z:29.905000686645508
- atom: CB x: -15.355999946594238 y:8.258999824523926 z:32.018001556396484
- atom: CG x: -16.48200035095215 y:8.942999839782715 z:31.2450008392334
- atom: CD1 x: -17.711000442504883 y:8.062999725341797 z:31.25
- atom: CD2 x: -16.80299949645996 y:10.270000457763672 z:31.878000259399414
- residue ILE has 8 atoms
- atom: N x: -14.506999969482422 y:6.693999767303467 z:28.940000534057617
- atom: CA x: -14.029000282287598 y:7.00600004196167 z:27.59600067138672
- atom: C x: -15.149999618530273 y:7.5229997634887695 z:26.711000442504883
- atom: O x: -16.299999237060547 y:7.142000198364258 z:26.871999740600586
- atom: CB x: -13.437999725341797 y:5.7779998779296875 z:26.885000228881836
- atom: CG1 x: -14.513999938964844 y:4.703999996185303 z:26.7549991607666
- atom: CG2 x: -12.199999809265137 y:5.271999835968018 z:27.624000549316406
- atom: CD1 x: -14.071999549865723 y:3.513000011444092 z:25.937999725341797
- residue PRO has 7 atoms
- atom: N x: -14.821000099182129 y:8.404000282287598 z:25.766000747680664
- atom: CA x: -15.810999870300293 y:8.979000091552734 z:24.83799934387207
- atom: C x: -16.490999221801758 y:7.840000152587891 z:24.062999725341797
- atom: O x: -15.810999870300293 y:6.901000022888184 z:23.624000549316406
- atom: CB x: -14.961999893188477 y:9.873000144958496 z:23.937999725341797
- atom: CG x: -13.824999809265137 y:10.289999961853027 z:24.843000411987305
- atom: CD x: -13.48900032043457 y:8.99899959564209 z:25.56800079345703
- residue LYS has 9 atoms
- atom: N x: -17.80900001525879 y:7.921999931335449 z:23.8799991607666
- atom: CA x: -18.554000854492188 y:6.8480000495910645 z:23.209999084472656
- atom: C x: -18.02199935913086 y:6.297999858856201 z:21.886999130249023
- atom: O x: -17.986000061035156 y:5.081999778747559 z:21.684999465942383
- atom: CB x: -20.007999420166016 y:7.251999855041504 z:22.993000030517578
- atom: CG x: -20.899999618530273 y:6.093999862670898 z:22.548999786376953
- atom: CD x: -21.552000045776367 y:5.38100004196167 z:23.746000289916992
- atom: CE x: -22.51300048828125 y:4.2829999923706055 z:23.280000686645508
- atom: NZ x: -23.47800064086914 y:3.8610000610351562 z:24.336000442504883
- residue ASP has 8 atoms
- atom: N x: -17.625999450683594 y:7.181000232696533 z:20.983999252319336
- atom: CA x: -17.1200008392334 y:6.754000186920166 z:19.68600082397461
- atom: C x: -15.680000305175781 y:6.251999855041504 z:19.711999893188477
- atom: O x: -15.003000259399414 y:6.2179999351501465 z:18.670000076293945
- atom: CB x: -17.22599983215332 y:7.90500020980835 z:18.687000274658203
- atom: CG x: -16.69099998474121 y:9.199000358581543 z:19.2450008392334
- atom: OD1 x: -15.991000175476074 y:9.913999557495117 z:18.503999710083008
- atom: OD2 x: -16.976999282836914 y:9.498000144958496 z:20.422000885009766
- residue GLN has 9 atoms
- atom: N x: -15.208000183105469 y:5.869999885559082 z:20.895000457763672
- atom: CA x: -13.840999603271484 y:5.36899995803833 z:21.02899932861328
- atom: C x: -13.864999771118164 y:4.067999839782715 z:21.81100082397461
- atom: O x: -12.859999656677246 y:3.375 z:21.93899917602539
- atom: CB x: -12.97700023651123 y:6.406000137329102 z:21.72800064086914
- atom: CG x: -13.196000099182129 y:7.809000015258789 z:21.20599937438965
- atom: CD x: -11.937000274658203 y:8.630999565124512 z:21.261999130249023
- atom: OE1 x: -11.22599983215332 y:8.621999740600586 z:22.266000747680664
- atom: NE2 x: -11.645999908447266 y:9.350000381469727 z:20.18199920654297
- residue TYR has 12 atoms
- atom: N x: -15.048999786376953 y:3.755000114440918 z:22.316999435424805
- atom: CA x: -15.3100004196167 y:2.546999931335449 z:23.077999114990234
- atom: C x: -14.543000221252441 y:1.3359999656677246 z:22.562999725341797
- atom: O x: -13.607999801635742 y:0.8619999885559082 z:23.190000534057617
- atom: CB x: -16.804000854492188 y:2.242000102996826 z:23.031999588012695
- atom: CG x: -17.19499969482422 y:1.00600004196167 z:23.79199981689453
- atom: CD1 x: -17.104999542236328 y:0.9660000205039978 z:25.180999755859375
- atom: CD2 x: -17.6299991607666 y:-0.13699999451637268 z:23.121000289916992
- atom: CE1 x: -17.437999725341797 y:-0.1850000023841858 z:25.889999389648438
- atom: CE2 x: -17.964000701904297 y:-1.2970000505447388 z:23.815000534057617
- atom: CZ x: -17.865999221801758 y:-1.319000005722046 z:25.20199966430664
- atom: OH x: -18.17799949645996 y:-2.4670000076293945 z:25.902000427246094
- residue TYR has 12 atoms
- atom: N x: -14.942999839782715 y:0.8270000219345093 z:21.410999298095703
- atom: CA x: -14.288000106811523 y:-0.3409999907016754 z:20.8700008392334
- atom: C x: -12.777999877929688 y:-0.21400000154972076 z:20.922000885009766
- atom: O x: -12.097000122070312 y:-1.1469999551773071 z:21.32200050354004
- atom: CB x: -14.779999732971191 y:-0.5960000157356262 z:19.445999145507812
- atom: CG x: -16.284000396728516 y:-0.7670000195503235 z:19.385000228881836
- atom: CD1 x: -16.902000427246094 y:-1.9190000295639038 z:19.878999710083008
- atom: CD2 x: -17.097999572753906 y:0.25699999928474426 z:18.899999618530273
- atom: CE1 x: -18.301000595092773 y:-2.0450000762939453 z:19.89900016784668
- atom: CE2 x: -18.493999481201172 y:0.1469999998807907 z:18.916000366210938
- atom: CZ x: -19.089000701904297 y:-1.0019999742507935 z:19.420000076293945
- atom: OH x: -20.4689998626709 y:-1.069000005722046 z:19.485000610351562
- residue CYS has 6 atoms
- atom: N x: -12.243000030517578 y:0.9359999895095825 z:20.545000076293945
- atom: CA x: -10.789999961853027 y:1.1050000190734863 z:20.573999404907227
- atom: C x: -10.241999626159668 y:1.0920000076293945 z:22.003999710083008
- atom: O x: -9.145000457763672 y:0.6069999933242798 z:22.257999420166016
- atom: CB x: -10.390999794006348 y:2.4019999504089355 z:19.875999450683594
- atom: SG x: -9.300999641418457 y:2.1089999675750732 z:18.48900032043457
- residue GLY has 4 atoms
- atom: N x: -11.019000053405762 y:1.6349999904632568 z:22.93199920654297
- atom: CA x: -10.604000091552734 y:1.6460000276565552 z:24.31800079345703
- atom: C x: -10.835000038146973 y:0.289000004529953 z:24.96299934387207
- atom: O x: -10.050999641418457 y:-0.16200000047683716 z:25.792999267578125
- residue VAL has 7 atoms
- atom: N x: -11.920000076293945 y:-0.36800000071525574 z:24.582000732421875
- atom: CA x: -12.220999717712402 y:-1.6660000085830688 z:25.134000778198242
- atom: C x: -11.131999969482422 y:-2.628000020980835 z:24.66900062561035
- atom: O x: -10.866999626159668 y:-3.63700008392334 z:25.305999755859375
- atom: CB x: -13.607000350952148 y:-2.1589999198913574 z:24.66699981689453
- atom: CG1 x: -13.576000213623047 y:-2.502000093460083 z:23.184999465942383
- atom: CG2 x: -14.036999702453613 y:-3.3519999980926514 z:25.479999542236328
- residue LEU has 8 atoms
- atom: N x: -10.487000465393066 y:-2.309999942779541 z:23.559999465942383
- atom: CA x: -9.4399995803833 y:-3.1710000038146973 z:23.065000534057617
- atom: C x: -8.232999801635742 y:-2.9670000076293945 z:23.94700050354004
- atom: O x: -7.6620001792907715 y:-3.9210000038146973 z:24.485000610351562
- atom: CB x: -9.074000358581543 y:-2.805999994277954 z:21.634000778198242
- atom: CG x: -7.961999893188477 y:-3.6480000019073486 z:21.007999420166016
- atom: CD1 x: -8.39900016784668 y:-5.107999801635742 z:20.95599937438965
- atom: CD2 x: -7.644999980926514 y:-3.119999885559082 z:19.613000869750977
- residue TYR has 12 atoms
- atom: N x: -7.855000019073486 y:-1.7059999704360962 z:24.0939998626709
- atom: CA x: -6.703000068664551 y:-1.3450000286102295 z:24.893999099731445
- atom: C x: -6.7870001792907715 y:-1.8880000114440918 z:26.312999725341797
- atom: O x: -5.877999782562256 y:-2.578000068664551 z:26.77199935913086
- atom: CB x: -6.550000190734863 y:0.17399999499320984 z:24.950000762939453
- atom: CG x: -5.408999919891357 y:0.6000000238418579 z:25.836000442504883
- atom: CD1 x: -4.098999977111816 y:0.5410000085830688 z:25.382999420166016
- atom: CD2 x: -5.636000156402588 y:0.9879999756813049 z:27.158000946044922
- atom: CE1 x: -3.0409998893737793 y:0.8510000109672546 z:26.216999053955078
- atom: CE2 x: -4.584000110626221 y:1.2960000038146973 z:28.003999710083008
- atom: CZ x: -3.2880001068115234 y:1.222000002861023 z:27.525999069213867
- atom: OH x: -2.2260000705718994 y:1.49399995803833 z:28.360000610351562
- residue PHE has 11 atoms
- atom: N x: -7.882999897003174 y:-1.565000057220459 z:26.993999481201172
- atom: CA x: -8.121000289916992 y:-1.968999981880188 z:28.378000259399414
- atom: C x: -8.307999610900879 y:-3.4790000915527344 z:28.600000381469727
- atom: O x: -8.409000396728516 y:-3.937999963760376 z:29.736000061035156
- atom: CB x: -9.335000038146973 y:-1.2009999752044678 z:28.913999557495117
- atom: CG x: -9.093999862670898 y:-0.5210000276565552 z:30.23200035095215
- atom: CD1 x: -7.830999851226807 y:-0.009999999776482582 z:30.555999755859375
- atom: CD2 x: -10.133000373840332 y:-0.37299999594688416 z:31.152999877929688
- atom: CE1 x: -7.607999801635742 y:0.6340000033378601 z:31.783000946044922
- atom: CE2 x: -9.920999526977539 y:0.27399998903274536 z:32.38800048828125
- atom: CZ x: -8.659000396728516 y:0.7749999761581421 z:32.70100021362305
- residue THR has 7 atoms
- atom: N x: -8.368000030517578 y:-4.252999782562256 z:27.52199935913086
- atom: CA x: -8.527000427246094 y:-5.695000171661377 z:27.659000396728516
- atom: C x: -7.127999782562256 y:-6.291999816894531 z:27.558000564575195
- atom: O x: -6.834000110626221 y:-7.348999977111816 z:28.125
- atom: CB x: -9.447999954223633 y:-6.2870001792907715 z:26.538000106811523
- atom: OG1 x: -10.755999565124512 y:-5.709000110626221 z:26.632999420166016
- atom: CG2 x: -9.590999603271484 y:-7.790999889373779 z:26.69499969482422
- residue GLY has 4 atoms
- atom: N x: -6.26200008392334 y:-5.583000183105469 z:26.841999053955078
- atom: CA x: -4.89900016784668 y:-6.0329999923706055 z:26.658000946044922
- atom: C x: -4.8379998207092285 y:-7.366000175476074 z:25.944000244140625
- atom: O x: -5.651000022888184 y:-7.636000156402588 z:25.055999755859375
- residue SER has 6 atoms
- atom: N x: -3.8940000534057617 y:-8.215999603271484 z:26.344999313354492
- atom: CA x: -2.950000047683716 y:-7.925000190734863 z:27.43899917602539
- atom: C x: -1.9830000400543213 y:-6.807000160217285 z:27.10099983215332
- atom: O x: -1.75600004196167 y:-6.502999782562256 z:25.93000030517578
- atom: CB x: -2.1080000400543213 y:-9.154000282287598 z:27.775999069213867
- atom: OG x: -1.0529999732971191 y:-9.291999816894531 z:26.840999603271484
- residue ASP has 8 atoms
- atom: N x: -1.3969999551773071 y:-6.228000164031982 z:28.145000457763672
- atom: CA x: -0.42899999022483826 y:-5.145999908447266 z:28.013999938964844
- atom: C x: 0.6370000243186951 y:-5.468999862670898 z:26.96299934387207
- atom: O x: 0.9330000281333923 y:-4.64900016784668 z:26.097000122070312
- atom: CB x: 0.22100000083446503 y:-4.900000095367432 z:29.3700008392334
- atom: CG x: 0.7870000004768372 y:-6.170000076293945 z:29.974000930786133
- atom: OD1 x: 1.9320000410079956 y:-6.541999816894531 z:29.61400032043457
- atom: OD2 x: 0.07400000095367432 y:-6.804999828338623 z:30.790000915527344
- residue ILE has 8 atoms
- atom: N x: 1.215999960899353 y:-6.6620001792907715 z:27.04199981689453
- atom: CA x: 2.2239999771118164 y:-7.057000160217285 z:26.06999969482422
- atom: C x: 1.5240000486373901 y:-7.189000129699707 z:24.72800064086914
- atom: O x: 2.115000009536743 y:-6.8979997634887695 z:23.695999145507812
- atom: CB x: 2.88700008392334 y:-8.416000366210938 z:26.416000366210938
- atom: CG1 x: 3.5199999809265137 y:-8.373000144958496 z:27.809999465942383
- atom: CG2 x: 3.9570000171661377 y:-8.734999656677246 z:25.385000228881836
- atom: CD1 x: 4.730000019073486 y:-7.460999965667725 z:27.913000106811523
- residue PHE has 11 atoms
- atom: N x: 0.27000001072883606 y:-7.638999938964844 z:24.740999221801758
- atom: CA x: -0.4909999966621399 y:-7.76800012588501 z:23.500999450683594
- atom: C x: -0.550000011920929 y:-6.389999866485596 z:22.841999053955078
- atom: O x: -0.3019999861717224 y:-6.252999782562256 z:21.641000747680664
- atom: CB x: -1.9140000343322754 y:-8.26200008392334 z:23.777000427246094
- atom: CG x: -2.802999973297119 y:-8.269000053405762 z:22.55900001525879
- atom: CD1 x: -4.051000118255615 y:-7.645999908447266 z:22.586000442504883
- atom: CD2 x: -2.3959999084472656 y:-8.892999649047852 z:21.381999969482422
- atom: CE1 x: -4.880000114440918 y:-7.64300012588501 z:21.461000442504883
- atom: CE2 x: -3.2209999561309814 y:-8.895000457763672 z:20.25200080871582
- atom: CZ x: -4.4629998207092285 y:-8.267000198364258 z:20.29599952697754
- residue ASN has 8 atoms
- atom: N x: -0.8730000257492065 y:-5.379000186920166 z:23.64900016784668
- atom: CA x: -0.9649999737739563 y:-3.994999885559082 z:23.198999404907227
- atom: C x: 0.36899998784065247 y:-3.4709999561309814 z:22.68199920654297
- atom: O x: 0.4090000092983246 y:-2.625 z:21.791000366210938
- atom: CB x: -1.4759999513626099 y:-3.121000051498413 z:24.334999084472656
- atom: CG x: -2.9230000972747803 y:-3.3919999599456787 z:24.648000717163086
- atom: OD1 x: -3.4649999141693115 y:-4.428999900817871 z:24.25200080871582
- atom: ND2 x: -3.565000057220459 y:-2.4709999561309814 z:25.361000061035156
- residue LYS has 9 atoms
- atom: N x: 1.4589999914169312 y:-3.9660000801086426 z:23.25200080871582
- atom: CA x: 2.7829999923706055 y:-3.575000047683716 z:22.798999786376953
- atom: C x: 2.8970000743865967 y:-4.184000015258789 z:21.39900016784668
- atom: O x: 3.390000104904175 y:-3.553999900817871 z:20.46500015258789
- atom: CB x: 3.865000009536743 y:-4.192999839782715 z:23.691999435424805
- atom: CG x: 4.803999900817871 y:-3.2190001010894775 z:24.374000549316406
- atom: CD x: 4.2220001220703125 y:-2.7219998836517334 z:25.68199920654297
- atom: CE x: 5.249000072479248 y:-1.906999945640564 z:26.447999954223633
- atom: NZ x: 4.71999979019165 y:-1.440000057220459 z:27.759000778198242
- residue ASN has 8 atoms
- atom: N x: 2.4240000247955322 y:-5.421000003814697 z:21.270999908447266
- atom: CA x: 2.4730000495910645 y:-6.144999980926514 z:20.006000518798828
- atom: C x: 1.680999994277954 y:-5.458000183105469 z:18.91900062561035
- atom: O x: 2.183000087738037 y:-5.236999988555908 z:17.82200050354004
- atom: CB x: 1.9279999732971191 y:-7.566999912261963 z:20.170000076293945
- atom: CG x: 2.819000005722046 y:-8.437000274658203 z:21.020999908447266
- atom: OD1 x: 4.0329999923706055 y:-8.473999977111816 z:20.822999954223633
- atom: ND2 x: 2.2230000495910645 y:-9.152000427246094 z:21.969999313354492
- residue MET has 8 atoms
- atom: N x: 0.43299999833106995 y:-5.135000228881836 z:19.23699951171875
- atom: CA x: -0.46299999952316284 y:-4.486000061035156 z:18.29400062561035
- atom: C x: -0.013000000268220901 y:-3.0759999752044678 z:17.910999298095703
- atom: O x: 0.13500000536441803 y:-2.7690000534057617 z:16.731000900268555
- atom: CB x: -1.8730000257492065 y:-4.451000213623047 z:18.875
- atom: CG x: -2.9140000343322754 y:-3.936000108718872 z:17.91699981689453
- atom: SD x: -4.511000156402588 y:-3.989000082015991 z:18.68199920654297
- atom: CE x: -5.236000061035156 y:-5.382999897003174 z:17.812000274658203
- residue ARG has 11 atoms
- atom: N x: 0.20499999821186066 y:-2.2179999351501465 z:18.899999618530273
- atom: CA x: 0.6489999890327454 y:-0.8560000061988831 z:18.618000030517578
- atom: C x: 2.0329999923706055 y:-0.8849999904632568 z:17.986000061035156
- atom: O x: 2.75600004196167 y:0.10899999737739563 z:18.027000427246094
- atom: CB x: 0.7039999961853027 y:-0.02199999988079071 z:19.905000686645508
- atom: CG x: -0.6389999985694885 y:0.16200000047683716 z:20.607999801635742
- atom: CD x: -0.546999990940094 y:1.11899995803833 z:21.801000595092773
- atom: NE x: 0.36800000071525574 y:0.6430000066757202 z:22.839000701904297
- atom: CZ x: 1.690000057220459 y:0.8040000200271606 z:22.823999404907227
- atom: NH1 x: 2.2869999408721924 y:1.440999984741211 z:21.822999954223633
- atom: NH2 x: 2.4210000038146973 y:0.3109999895095825 z:23.812000274658203
- residue ALA has 5 atoms
- atom: N x: 2.3959999084472656 y:-2.0299999713897705 z:17.41200065612793
- atom: CA x: 3.694999933242798 y:-2.2079999446868896 z:16.777000427246094
- atom: C x: 3.558000087738037 y:-2.7639999389648438 z:15.357999801635742
- atom: O x: 4.267000198364258 y:-2.3389999866485596 z:14.446999549865723
- atom: CB x: 4.560999870300293 y:-3.131999969482422 z:17.625
- residue HIS has 10 atoms
- atom: N x: 2.6530001163482666 y:-3.7170000076293945 z:15.168000221252441
- atom: CA x: 2.447000026702881 y:-4.2870001792907715 z:13.842000007629395
- atom: C x: 1.50600004196167 y:-3.38100004196167 z:13.050000190734863
- atom: O x: 1.3990000486373901 y:-3.496000051498413 z:11.829999923706055
- atom: CB x: 1.8489999771118164 y:-5.692999839782715 z:13.930999755859375
- atom: CG x: 2.2960000038146973 y:-6.5980000495910645 z:12.826000213623047
- atom: ND1 x: 3.5880000591278076 y:-7.076000213623047 z:12.737000465393066
- atom: CD2 x: 1.6419999599456787 y:-7.076000213623047 z:11.741000175476074
- atom: CE1 x: 3.7090001106262207 y:-7.809999942779541 z:11.645000457763672
- atom: NE2 x: 2.5429999828338623 y:-7.827000141143799 z:11.02299976348877
- residue ALA has 5 atoms
- atom: N x: 0.828000009059906 y:-2.4809999465942383 z:13.76200008392334
- atom: CA x: -0.10000000149011612 y:-1.5369999408721924 z:13.149999618530273
- atom: C x: 0.6909999847412109 y:-0.40799999237060547 z:12.517999649047852
- atom: O x: 0.12399999797344208 y:0.5239999890327454 z:11.958000183105469
- atom: CB x: -1.0499999523162842 y:-0.9750000238418579 z:14.199999809265137
- residue LEU has 8 atoms
- atom: N x: 2.009999990463257 y:-0.4959999918937683 z:12.625
- atom: CA x: 2.8910000324249268 y:0.5099999904632568 z:12.059000015258789
- atom: C x: 3.50600004196167 y:-0.03500000014901161 z:10.78499984741211
- atom: O x: 3.7899999618530273 y:0.7179999947547913 z:9.859999656677246
- atom: CB x: 3.9809999465942383 y:0.875 z:13.062000274658203
- atom: CG x: 3.4670000076293945 y:1.5980000495910645 z:14.305000305175781
- atom: CD1 x: 4.455999851226807 y:1.4429999589920044 z:15.440999984741211
- atom: CD2 x: 3.243000030517578 y:3.062999963760376 z:13.977999687194824
- residue GLU has 9 atoms
- atom: N x: 3.7119998931884766 y:-1.3480000495910645 z:10.744999885559082
- atom: CA x: 4.270999908447266 y:-2.0 z:9.562999725341797
- atom: C x: 3.1630001068115234 y:-2.0859999656677246 z:8.519000053405762
- atom: O x: 3.4089999198913574 y:-2.381999969482422 z:7.3460001945495605
- atom: CB x: 4.775000095367432 y:-3.4110000133514404 z:9.897000312805176
- atom: CG x: 6.122000217437744 y:-3.4679999351501465 z:10.611000061035156
- atom: CD x: 6.538000106811523 y:-4.894000053405762 z:10.965999603271484
- atom: OE1 x: 5.827000141143799 y:-5.545000076293945 z:11.767999649047852
- atom: OE2 x: 7.573999881744385 y:-5.364999771118164 z:10.442000389099121
- residue LYS has 9 atoms
- atom: N x: 1.937000036239624 y:-1.8250000476837158 z:8.96399974822998
- atom: CA x: 0.7760000228881836 y:-1.8489999771118164 z:8.09000015258789
- atom: C x: 0.4650000035762787 y:-0.4359999895095825 z:7.625
- atom: O x: -0.1469999998807907 y:-0.23800000548362732 z:6.577000141143799
- atom: CB x: -0.42500001192092896 y:-2.430999994277954 z:8.829000473022461
- atom: CG x: -0.34700000286102295 y:-3.931999921798706 z:9.03600025177002
- atom: CD x: -0.3109999895095825 y:-4.669000148773193 z:7.701000213623047
- atom: CE x: -0.36899998784065247 y:-6.182000160217285 z:7.88100004196167
- atom: NZ x: -0.4009999930858612 y:-6.886000156402588 z:6.570000171661377
- residue GLY has 4 atoms
- atom: N x: 0.8970000147819519 y:0.5450000166893005 z:8.40999984741211
- atom: CA x: 0.6600000262260437 y:1.930999994277954 z:8.053999900817871
- atom: C x: -0.44699999690055847 y:2.5869998931884766 z:8.862000465393066
- atom: O x: -1.2719999551773071 y:3.318000078201294 z:8.3100004196167
- residue PHE has 11 atoms
- atom: N x: -0.4729999899864197 y:2.3239998817443848 z:10.166999816894531
- atom: CA x: -1.4789999723434448 y:2.8940000534057617 z:11.060999870300293
- atom: C x: -0.8289999961853027 y:3.25 z:12.390000343322754
- atom: O x: 0.3659999966621399 y:3.0260000228881836 z:12.576000213623047
- atom: CB x: -2.614000082015991 y:1.8949999809265137 z:11.293999671936035
- atom: CG x: -3.365000009536743 y:1.534999966621399 z:10.04699993133545
- atom: CD1 x: -2.7880001068115234 y:0.7170000076293945 z:9.079999923706055
- atom: CD2 x: -4.638999938964844 y:2.0510001182556152 z:9.812999725341797
- atom: CE1 x: -3.4639999866485596 y:0.42100000381469727 z:7.896999835968018
- atom: CE2 x: -5.323999881744385 y:1.7610000371932983 z:8.633999824523926
- atom: CZ x: -4.734000205993652 y:0.9449999928474426 z:7.675000190734863
- residue THR has 7 atoms
- atom: N x: -1.6059999465942383 y:3.812999963760376 z:13.309000015258789
- atom: CA x: -1.0820000171661377 y:4.171000003814697 z:14.621000289916992
- atom: C x: -2.1449999809265137 y:4.130000114440918 z:15.713000297546387
- atom: O x: -2.8350000381469727 y:5.118000030517578 z:15.982999801635742
- atom: CB x: -0.4440000057220459 y:5.560999870300293 z:14.616999626159668
- atom: OG1 x: -0.1979999989271164 y:5.96999979019165 z:15.970999717712402
- atom: CG2 x: -1.3569999933242798 y:6.568999767303467 z:13.937999725341797
- residue ILE has 8 atoms
- atom: N x: -2.260999917984009 y:2.9660000801086426 z:16.34000015258789
- atom: CA x: -3.2179999351501465 y:2.749000072479248 z:17.410999298095703
- atom: C x: -2.7730000019073486 y:3.4049999713897705 z:18.715999603271484
- atom: O x: -1.5839999914169312 y:3.5250000953674316 z:18.99799919128418
- atom: CB x: -3.384000062942505 y:1.25 z:17.70800018310547
- atom: CG1 x: -3.8320000171661377 y:0.5090000033378601 z:16.45400047302246
- atom: CG2 x: -4.36899995803833 y:1.0549999475479126 z:18.841999053955078
- atom: CD1 x: -3.9230000972747803 y:-0.9929999709129333 z:16.635000228881836
- residue ASN has 8 atoms
- atom: N x: -3.750999927520752 y:3.8329999446868896 z:19.49799919128418
- atom: CA x: -3.5160000324249268 y:4.416999816894531 z:20.804000854492188
- atom: C x: -4.771999835968018 y:3.9769999980926514 z:21.53499984741211
- atom: O x: -5.671000003814697 y:3.4179999828338623 z:20.91699981689453
- atom: CB x: -3.364000082015991 y:5.951000213623047 z:20.72800064086914
- atom: CG x: -4.677000045776367 y:6.679999828338623 z:20.570999145507812
- atom: OD1 x: -5.383999824523926 y:6.507999897003174 z:19.586000442504883
- atom: ND2 x: -5.002999782562256 y:7.520999908447266 z:21.547000885009766
- residue GLU has 9 atoms
- atom: N x: -4.8480000495910645 y:4.176000118255615 z:22.83799934387207
- atom: CA x: -6.034999847412109 y:3.7279999256134033 z:23.54800033569336
- atom: C x: -7.315999984741211 y:4.426000118255615 z:23.10700035095215
- atom: O x: -8.413999557495117 y:4.002999782562256 z:23.4689998626709
- atom: CB x: -5.839000225067139 y:3.9059998989105225 z:25.049999237060547
- atom: CG x: -5.285999774932861 y:5.250999927520752 z:25.452999114990234
- atom: CD x: -5.427000045776367 y:5.492000102996826 z:26.93899917602539
- atom: OE1 x: -6.584000110626221 y:5.568999767303467 z:27.41699981689453
- atom: OE2 x: -4.386000156402588 y:5.5960001945495605 z:27.628999710083008
- residue TYR has 12 atoms
- atom: N x: -7.186999797821045 y:5.4730000495910645 z:22.301000595092773
- atom: CA x: -8.359000205993652 y:6.21999979019165 z:21.871999740600586
- atom: C x: -8.829000473022461 y:6.129000186920166 z:20.430999755859375
- atom: O x: -10.012999534606934 y:6.314000129699707 z:20.163000106811523
- atom: CB x: -8.189000129699707 y:7.686999797821045 z:22.2549991607666
- atom: CG x: -8.407999992370605 y:7.910999774932861 z:23.726999282836914
- atom: CD1 x: -9.579999923706055 y:7.461999893188477 z:24.343000411987305
- atom: CD2 x: -7.442999839782715 y:8.538999557495117 z:24.511999130249023
- atom: CE1 x: -9.788999557495117 y:7.626999855041504 z:25.70400047302246
- atom: CE2 x: -7.639999866485596 y:8.711000442504883 z:25.878999710083008
- atom: CZ x: -8.817999839782715 y:8.24899959564209 z:26.469999313354492
- atom: OH x: -9.015999794006348 y:8.385000228881836 z:27.82900047302246
- residue THR has 7 atoms
- atom: N x: -7.926000118255615 y:5.84499979019165 z:19.503999710083008
- atom: CA x: -8.315999984741211 y:5.76200008392334 z:18.10300064086914
- atom: C x: -7.355000019073486 y:4.928999900817871 z:17.274999618530273
- atom: O x: -6.382999897003174 y:4.380000114440918 z:17.781999588012695
- atom: CB x: -8.359000205993652 y:7.159999847412109 z:17.450000762939453
- atom: OG1 x: -7.072000026702881 y:7.771999835968018 z:17.58099937438965
- atom: CG2 x: -9.40999984741211 y:8.045999526977539 z:18.10300064086914
- residue ILE has 8 atoms
- atom: N x: -7.651000022888184 y:4.840000152587891 z:15.984999656677246
- atom: CA x: -6.809999942779541 y:4.129000186920166 z:15.03600025177002
- atom: C x: -6.605000019073486 y:5.14300012588501 z:13.92300033569336
- atom: O x: -7.561999797821045 y:5.546000003814697 z:13.265999794006348
- atom: CB x: -7.505000114440918 y:2.8489999771118164 z:14.496000289916992
- atom: CG1 x: -6.677000045776367 y:2.243000030517578 z:13.366000175476074
- atom: CG2 x: -8.907999992370605 y:3.1640000343322754 z:14.024999618530273
- atom: CD1 x: -5.2870001792907715 y:1.8519999980926514 z:13.770000457763672
- residue ARG has 11 atoms
- atom: N x: -5.361999988555908 y:5.576000213623047 z:13.73900032043457
- atom: CA x: -5.0320000648498535 y:6.578000068664551 z:12.72599983215332
- atom: C x: -4.03000020980835 y:6.0970001220703125 z:11.682000160217285
- atom: O x: -3.0840001106262207 y:5.367000102996826 z:11.996000289916992
- atom: CB x: -4.446000099182129 y:7.829999923706055 z:13.387999534606934
- atom: CG x: -5.349999904632568 y:8.541000366210938 z:14.371999740600586
- atom: CD x: -4.564000129699707 y:9.571000099182129 z:15.16100025177002
- atom: NE x: -5.436999797821045 y:10.416000366210938 z:15.961999893188477
- atom: CZ x: -6.255000114440918 y:11.333999633789062 z:15.456000328063965
- atom: NH1 x: -6.309000015258789 y:11.53499984741211 z:14.142999649047852
- atom: NH2 x: -7.034999847412109 y:12.043000221252441 z:16.260000228881836
- residue PRO has 7 atoms
- atom: N x: -4.21999979019165 y:6.513000011444092 z:10.418999671936035
- atom: CA x: -3.3289999961853027 y:6.136000156402588 z:9.314000129699707
- atom: C x: -2.0810000896453857 y:7.013000011444092 z:9.279999732971191
- atom: O x: -2.1059999465942383 y:8.13700008392334 z:9.77299976348877
- atom: CB x: -4.203000068664551 y:6.354000091552734 z:8.08899974822998
- atom: CG x: -5.043000221252441 y:7.53000020980835 z:8.503999710083008
- atom: CD x: -5.443999767303467 y:7.158999919891357 z:9.90999984741211
- residue LEU has 8 atoms
- atom: N x: -0.9909999966621399 y:6.5 z:8.71500015258789
- atom: CA x: 0.23899999260902405 y:7.2820000648498535 z:8.612000465393066
- atom: C x: 0.4749999940395355 y:7.710999965667725 z:7.160999774932861
- atom: O x: 0.6140000224113464 y:6.875999927520752 z:6.260000228881836
- atom: CB x: 1.4550000429153442 y:6.493000030517578 z:9.128000259399414
- atom: CG x: 1.6330000162124634 y:6.239999771118164 z:10.633000373840332
- atom: CD1 x: 3.072000026702881 y:5.822000026702881 z:10.888999938964844
- atom: CD2 x: 1.3270000219345093 y:7.493000030517578 z:11.430999755859375
- residue GLY has 4 atoms
- atom: N x: 0.5199999809265137 y:9.02400016784668 z:6.947000026702881
- atom: CA x: 0.7110000252723694 y:9.560999870300293 z:5.611999988555908
- atom: C x: 2.055000066757202 y:9.293999671936035 z:4.956999778747559
- atom: O x: 2.5769999027252197 y:8.173999786376953 z:5.01200008392334
- residue VAL has 5 atoms
- atom: N x: 2.6010000705718994 y:10.335000038146973 z:4.326000213623047
- atom: CA x: 3.885999917984009 y:10.26200008392334 z:3.632999897003174
- atom: C x: 5.041999816894531 y:10.295999526977539 z:4.625999927520752
- atom: O x: 5.809000015258789 y:9.336999893188477 z:4.73799991607666
- atom: CB x: 4.046000003814697 y:11.435999870300293 z:2.6449999809265137
- residue THR has 7 atoms
- atom: N x: 5.1620001792907715 y:11.411999702453613 z:5.3379998207092285
- atom: CA x: 6.205999851226807 y:11.58899974822998 z:6.3429999351501465
- atom: C x: 5.745999813079834 y:10.847000122070312 z:7.5879998207092285
- atom: O x: 6.538000106811523 y:10.524999618530273 z:8.479999542236328
- atom: CB x: 6.401000022888184 y:13.09000015258789 z:6.693999767303467
- atom: OG1 x: 7.418000221252441 y:13.229000091552734 z:7.696000099182129
- atom: CG2 x: 5.10099983215332 y:13.690999984741211 z:7.216000080108643
- residue GLY has 4 atoms
- atom: N x: 4.451000213623047 y:10.559000015258789 z:7.620999813079834
- atom: CA x: 3.871999979019165 y:9.878999710083008 z:8.756999969482422
- atom: C x: 3.1549999713897705 y:10.939000129699707 z:9.560999870300293
- atom: O x: 3.7009999752044678 y:11.47599983215332 z:10.526000022888184
- residue VAL has 7 atoms
- atom: N x: 1.9320000410079956 y:11.253999710083008 z:9.142000198364258
- atom: CA x: 1.121000051498413 y:12.267000198364258 z:9.810999870300293
- atom: C x: -0.13600000739097595 y:11.644000053405762 z:10.413000106811523
- atom: O x: -0.7139999866485596 y:10.715999603271484 z:9.848999977111816
- atom: CB x: 0.6930000185966492 y:13.37600040435791 z:8.821000099182129
- atom: CG1 x: 0.11800000071525574 y:14.564000129699707 z:9.583999633789062
- atom: CG2 x: 1.8819999694824219 y:13.795000076293945 z:7.960999965667725
- residue ALA has 5 atoms
- atom: N x: -0.5550000071525574 y:12.157999992370605 z:11.562000274658203
- atom: CA x: -1.746000051498413 y:11.645999908447266 z:12.223999977111816
- atom: C x: -2.989000082015991 y:11.880000114440918 z:11.366999626159668
- atom: O x: -3.7219998836517334 y:12.854999542236328 z:11.567999839782715
- atom: CB x: -1.9160000085830688 y:12.317000389099121 z:13.588000297546387
- residue GLY has 4 atoms
- atom: N x: -3.2219998836517334 y:10.994000434875488 z:10.402999877929688
- atom: CA x: -4.394999980926514 y:11.140999794006348 z:9.5649995803833
- atom: C x: -5.603000164031982 y:11.170000076293945 z:10.477999687194824
- atom: O x: -5.493000030517578 y:10.807000160217285 z:11.645999908447266
- residue GLU has 9 atoms
- atom: N x: -6.754000186920166 y:11.597999572753906 z:9.97599983215332
- atom: CA x: -7.928999900817871 y:11.63700008392334 z:10.82699966430664
- atom: C x: -8.236000061035156 y:10.243000030517578 z:11.357999801635742
- atom: O x: -7.811999797821045 y:9.243000030517578 z:10.781999588012695
- atom: CB x: -9.128000259399414 y:12.204000473022461 z:10.067999839782715
- atom: CG x: -8.973999977111816 y:13.675999641418457 z:9.729000091552734
- atom: CD x: -8.668999671936035 y:14.520999908447266 z:10.954000473022461
- atom: OE1 x: -9.545999526977539 y:14.63599967956543 z:11.836999893188477
- atom: OE2 x: -7.546000003814697 y:15.064000129699707 z:11.038999557495117
- residue PRO has 7 atoms
- atom: N x: -8.956999778747559 y:10.168000221252441 z:12.486000061035156
- atom: CA x: -9.35099983215332 y:8.928999900817871 z:13.156000137329102
- atom: C x: -10.333999633789062 y:8.079000473022461 z:12.36299991607666
- atom: O x: -11.458999633789062 y:8.48799991607666 z:12.119999885559082
- atom: CB x: -9.963000297546387 y:9.427000045776367 z:14.458000183105469
- atom: CG x: -9.27299976348877 y:10.722000122070312 z:14.6899995803833
- atom: CD x: -9.281999588012695 y:11.329000473022461 z:13.32800006866455
- residue LEU has 8 atoms
- atom: N x: -9.914999961853027 y:6.882999897003174 z:11.980999946594238
- atom: CA x: -10.781000137329102 y:5.994999885559082 z:11.220000267028809
- atom: C x: -12.0 y:5.538000106811523 z:12.052000045776367
- atom: O x: -11.925000190734863 y:5.4029998779296875 z:13.284000396728516
- atom: CB x: -9.970000267028809 y:4.785999774932861 z:10.727999687194824
- atom: CG x: -8.637999534606934 y:5.125 z:10.034000396728516
- atom: CD1 x: -7.88100004196167 y:3.8529999256134033 z:9.711999893188477
- atom: CD2 x: -8.88599967956543 y:5.926000118255615 z:8.770999908447266
- residue PRO has 7 atoms
- atom: N x: -13.142999649047852 y:5.301000118255615 z:11.381999969482422
- atom: CA x: -14.369999885559082 y:4.864999771118164 z:12.055999755859375
- atom: C x: -14.175000190734863 y:3.4600000381469727 z:12.61299991607666
- atom: O x: -13.482000350952148 y:2.6470000743865967 z:12.015000343322754
- atom: CB x: -15.402999877929688 y:4.925000190734863 z:10.942999839782715
- atom: CG x: -14.595000267028809 y:4.534999847412109 z:9.737000465393066
- atom: CD x: -13.314000129699707 y:5.303999900817871 z:9.918000221252441
- residue VAL has 7 atoms
- atom: N x: -14.791000366210938 y:3.1710000038146973 z:13.751999855041504
- atom: CA x: -14.619999885559082 y:1.871999979019165 z:14.383999824523926
- atom: C x: -15.878000259399414 y:1.4279999732971191 z:15.11400032043457
- atom: O x: -16.320999145507812 y:2.0989999771118164 z:16.045000076293945
- atom: CB x: -13.45199966430664 y:1.9279999732971191 z:15.414999961853027
- atom: CG1 x: -13.395000457763672 y:0.6629999876022339 z:16.229000091552734
- atom: CG2 x: -12.142000198364258 y:2.13700008392334 z:14.706000328063965
- residue ASP has 8 atoms
- atom: N x: -16.44499969482422 y:0.29499998688697815 z:14.708000183105469
- atom: CA x: -17.641000747680664 y:-0.2070000022649765 z:15.37600040435791
- atom: C x: -17.44300079345703 y:-1.562000036239624 z:16.04199981689453
- atom: O x: -18.38599967956543 y:-2.1410000324249268 z:16.584999084472656
- atom: CB x: -18.812000274658203 y:-0.27399998903274536 z:14.397000312805176
- atom: CG x: -19.378000259399414 y:1.090999960899353 z:14.090999603271484
- atom: OD1 x: -19.974000930786133 y:1.7089999914169312 z:15.01099967956543
- atom: OD2 x: -19.208999633789062 y:1.5449999570846558 z:12.937000274658203
- residue SER has 6 atoms
- atom: N x: -16.21299934387207 y:-2.056999921798706 z:16.011999130249023
- atom: CA x: -15.904000282287598 y:-3.3350000381469727 z:16.624000549316406
- atom: C x: -14.416999816894531 y:-3.440999984741211 z:16.860000610351562
- atom: O x: -13.630000114440918 y:-2.6700000762939453 z:16.297000885009766
- atom: CB x: -16.336000442504883 y:-4.482999801635742 z:15.71399974822998
- atom: OG x: -15.477999687194824 y:-4.5929999351501465 z:14.583999633789062
- residue GLU has 9 atoms
- atom: N x: -14.02299976348877 y:-4.389999866485596 z:17.70199966430664
- atom: CA x: -12.605999946594238 y:-4.583000183105469 z:17.94700050354004
- atom: C x: -12.114999771118164 y:-5.175000190734863 z:16.631000518798828
- atom: O x: -11.086999893188477 y:-4.754000186920166 z:16.094999313354492
- atom: CB x: -12.359999656677246 y:-5.572000026702881 z:19.093000411987305
- atom: CG x: -12.958000183105469 y:-5.177999973297119 z:20.433000564575195
- atom: CD x: -12.744000434875488 y:-6.241000175476074 z:21.503000259399414
- atom: OE1 x: -11.678000450134277 y:-6.239999771118164 z:22.149999618530273
- atom: OE2 x: -13.63700008392334 y:-7.091000080108643 z:21.690000534057617
- residue LYS has 9 atoms
- atom: N x: -12.881999969482422 y:-6.133999824523926 z:16.10300064086914
- atom: CA x: -12.550999641418457 y:-6.808000087738037 z:14.845000267028809
- atom: C x: -12.190999984741211 y:-5.770999908447266 z:13.795000076293945
- atom: O x: -11.251999855041504 y:-5.955999851226807 z:13.017000198364258
- atom: CB x: -13.72700023651123 y:-7.6579999923706055 z:14.350000381469727
- atom: CG x: -13.319999694824219 y:-8.717000007629395 z:13.312999725341797
- atom: CD x: -14.470999717712402 y:-9.666000366210938 z:12.951000213623047
- atom: CE x: -13.95300006866455 y:-10.96500015258789 z:12.319999694824219
- atom: NZ x: -13.071999549865723 y:-10.746999740600586 z:11.128999710083008
- residue ASP has 8 atoms
- atom: N x: -12.949999809265137 y:-4.681000232696533 z:13.784000396728516
- atom: CA x: -12.708999633789062 y:-3.5869998931884766 z:12.864999771118164
- atom: C x: -11.27299976348877 y:-3.0889999866485596 z:12.994000434875488
- atom: O x: -10.62399959564209 y:-2.819999933242798 z:11.98799991607666
- atom: CB x: -13.6850004196167 y:-2.4489998817443848 z:13.14900016784668
- atom: CG x: -15.019000053405762 y:-2.638000011444092 z:12.45199966430664
- atom: OD1 x: -15.416999816894531 y:-3.799999952316284 z:12.22700023651123
- atom: OD2 x: -15.673999786376953 y:-1.6230000257492065 z:12.137999534606934
- residue ILE has 8 atoms
- atom: N x: -10.767000198364258 y:-2.9700000286102295 z:14.220000267028809
- atom: CA x: -9.394000053405762 y:-2.506999969482422 z:14.414999961853027
- atom: C x: -8.388999938964844 y:-3.507999897003174 z:13.859999656677246
- atom: O x: -7.361000061035156 y:-3.1089999675750732 z:13.307999610900879
- atom: CB x: -9.074000358581543 y:-2.252000093460083 z:15.904000282287598
- atom: CG1 x: -9.920999526977539 y:-1.0920000076293945 z:16.42300033569336
- atom: CG2 x: -7.604000091552734 y:-1.9160000085830688 z:16.07200050354004
- atom: CD1 x: -9.934000015258789 y:-0.9750000238418579 z:17.92300033569336
- residue PHE has 11 atoms
- atom: N x: -8.67199993133545 y:-4.802999973297119 z:13.996999740600586
- atom: CA x: -7.755000114440918 y:-5.816999912261963 z:13.468000411987305
- atom: C x: -7.697000026702881 y:-5.741000175476074 z:11.954000473022461
- atom: O x: -6.613999843597412 y:-5.801000118255615 z:11.366000175476074
- atom: CB x: -8.1850004196167 y:-7.229000091552734 z:13.838000297546387
- atom: CG x: -8.031000137329102 y:-7.553999900817871 z:15.281000137329102
- atom: CD1 x: -8.97599983215332 y:-7.142000198364258 z:16.200000762939453
- atom: CD2 x: -6.9629998207092285 y:-8.329000473022461 z:15.71399974822998
- atom: CE1 x: -8.86400032043457 y:-7.50600004196167 z:17.531999588012695
- atom: CE2 x: -6.8420000076293945 y:-8.699000358581543 z:17.04400062561035
- atom: CZ x: -7.793000221252441 y:-8.289999961853027 z:17.95400047302246
- residue ASP has 8 atoms
- atom: N x: -8.868000030517578 y:-5.627999782562256 z:11.32800006866455
- atom: CA x: -8.956000328063965 y:-5.539999961853027 z:9.875
- atom: C x: -7.9770002365112305 y:-4.494999885559082 z:9.343000411987305
- atom: O x: -7.117000102996826 y:-4.798999786376953 z:8.513999938964844
- atom: CB x: -10.378999710083008 y:-5.177999973297119 z:9.451000213623047
- atom: CG x: -11.38599967956543 y:-6.243000030517578 z:9.824000358581543
- atom: OD1 x: -11.04800033569336 y:-7.438000202178955 z:9.717000007629395
- atom: OD2 x: -12.520000457763672 y:-5.888000011444092 z:10.206999778747559
- residue TYR has 12 atoms
- atom: N x: -8.109999656677246 y:-3.2669999599456787 z:9.835000038146973
- atom: CA x: -7.236000061035156 y:-2.171999931335449 z:9.430000305175781
- atom: C x: -5.750999927520752 y:-2.553999900817871 z:9.388999938964844
- atom: O x: -5.015999794006348 y:-2.0889999866485596 z:8.51200008392334
- atom: CB x: -7.4120001792907715 y:-0.9829999804496765 z:10.37600040435791
- atom: CG x: -8.711999893188477 y:-0.2290000021457672 z:10.222999572753906
- atom: CD1 x: -8.961999893188477 y:0.5600000023841858 z:9.097999572753906
- atom: CD2 x: -9.673999786376953 y:-0.26100000739097595 z:11.227999687194824
- atom: CE1 x: -10.13700008392334 y:1.3040000200271606 z:8.98799991607666
- atom: CE2 x: -10.85200023651123 y:0.4740000069141388 z:11.125
- atom: CZ x: -11.074000358581543 y:1.253999948501587 z:10.008000373840332
- atom: OH x: -12.22599983215332 y:1.99399995803833 z:9.934000015258789
- residue ILE has 8 atoms
- atom: N x: -5.306000232696533 y:-3.385999917984009 z:10.331999778747559
- atom: CA x: -3.9000000953674316 y:-3.7909998893737793 z:10.378999710083008
- atom: C x: -3.6470000743865967 y:-5.202000141143799 z:9.848999977111816
- atom: O x: -2.5460000038146973 y:-5.744999885559082 z:9.998000144958496
- atom: CB x: -3.3369998931884766 y:-3.7060000896453857 z:11.810999870300293
- atom: CG1 x: -4.114999771118164 y:-4.64900016784668 z:12.734999656677246
- atom: CG2 x: -3.4070000648498535 y:-2.2730000019073486 z:12.29800033569336
- atom: CD1 x: -3.5190000534057617 y:-4.806000232696533 z:14.109000205993652
- residue GLN has 9 atoms
- atom: N x: -4.672999858856201 y:-5.791999816894531 z:9.23799991607666
- atom: CA x: -4.566999912261963 y:-7.129000186920166 z:8.661999702453613
- atom: C x: -3.9079999923706055 y:-8.130999565124512 z:9.602999687194824
- atom: O x: -2.7799999713897705 y:-8.581999778747559 z:9.387999534606934
- atom: CB x: -3.802999973297119 y:-7.051000118255615 z:7.3420000076293945
- atom: CG x: -4.520999908447266 y:-6.198999881744385 z:6.308000087738037
- atom: CD x: -3.6029999256134033 y:-5.708000183105469 z:5.218999862670898
- atom: OE1 x: -2.9730000495910645 y:-6.502999782562256 z:4.520999908447266
- atom: NE2 x: -3.515000104904175 y:-4.388000011444092 z:5.065999984741211
- residue TRP has 14 atoms
- atom: N x: -4.6479997634887695 y:-8.468000411987305 z:10.651000022888184
- atom: CA x: -4.2129998207092285 y:-9.406999588012695 z:11.666000366210938
- atom: C x: -5.4029998779296875 y:-10.319000244140625 z:11.925000190734863
- atom: O x: -6.546999931335449 y:-9.857000350952148 z:11.963000297546387
- atom: CB x: -3.867000102996826 y:-8.654000282287598 z:12.949999809265137
- atom: CG x: -2.4119999408721924 y:-8.647000312805176 z:13.345000267028809
- atom: CD1 x: -1.3270000219345093 y:-8.75100040435791 z:12.519000053405762
- atom: CD2 x: -1.8910000324249268 y:-8.4350004196167 z:14.666000366210938
- atom: NE1 x: -0.164000004529953 y:-8.61299991607666 z:13.244999885559082
- atom: CE2 x: -0.4819999933242798 y:-8.416000366210938 z:14.562999725341797
- atom: CE3 x: -2.4809999465942383 y:-8.253999710083008 z:15.925999641418457
- atom: CZ2 x: 0.34700000286102295 y:-8.222999572753906 z:15.670999526977539
- atom: CZ3 x: -1.6579999923706055 y:-8.062000274658203 z:17.027000427246094
- atom: CH2 x: -0.257999986410141 y:-8.04800033569336 z:16.891000747680664
- residue LYS has 9 atoms
- atom: N x: -5.131999969482422 y:-11.611000061035156 z:12.079999923706055
- atom: CA x: -6.172999858856201 y:-12.593999862670898 z:12.36400032043457
- atom: C x: -6.895999908447266 y:-12.119000434875488 z:13.630999565124512
- atom: O x: -6.25 y:-11.840999603271484 z:14.640000343322754
- atom: CB x: -5.515999794006348 y:-13.961000442504883 z:12.612000465393066
- atom: CG x: -6.453999996185303 y:-15.081999778747559 z:13.057999610900879
- atom: CD x: -5.710000038146973 y:-16.18600082397461 z:13.833000183105469
- atom: CE x: -4.585000038146973 y:-16.8439998626709 z:13.029000282287598
- atom: NZ x: -3.36299991607666 y:-15.994999885559082 z:12.892999649047852
- residue TYR has 12 atoms
- atom: N x: -8.222000122070312 y:-12.010000228881836 z:13.583000183105469
- atom: CA x: -8.960000038146973 y:-11.565999984741211 z:14.760000228881836
- atom: C x: -8.760000228881836 y:-12.576000213623047 z:15.869999885559082
- atom: O x: -8.963000297546387 y:-13.774999618530273 z:15.677000045776367
- atom: CB x: -10.458000183105469 y:-11.418999671936035 z:14.470000267028809
- atom: CG x: -11.286999702453613 y:-11.11299991607666 z:15.708000183105469
- atom: CD1 x: -10.930999755859375 y:-10.07800006866455 z:16.576000213623047
- atom: CD2 x: -12.423999786376953 y:-11.857999801635742 z:16.016000747680664
- atom: CE1 x: -11.687000274658203 y:-9.791999816894531 z:17.722999572753906
- atom: CE2 x: -13.187000274658203 y:-11.57800006866455 z:17.160999298095703
- atom: CZ x: -12.8100004196167 y:-10.543999671936035 z:18.007999420166016
- atom: OH x: -13.557000160217285 y:-10.265999794006348 z:19.131999969482422
- residue ARG has 11 atoms
- atom: N x: -8.35099983215332 y:-12.07800006866455 z:17.030000686645508
- atom: CA x: -8.109999656677246 y:-12.918000221252441 z:18.19099998474121
- atom: C x: -9.045999526977539 y:-12.545000076293945 z:19.327999114990234
- atom: O x: -9.180000305175781 y:-11.371999740600586 z:19.68600082397461
- atom: CB x: -6.64300012588501 y:-12.793999671936035 z:18.628999710083008
- atom: CG x: -5.697000026702881 y:-13.623000144958496 z:17.770999908447266
- atom: CD x: -4.306000232696533 y:-13.03600025177002 z:17.694000244140625
- atom: NE x: -3.683000087738037 y:-12.902999877929688 z:19.003999710083008
- atom: CZ x: -2.4519999027252197 y:-12.432999610900879 z:19.19300079345703
- atom: NH1 x: -1.718999981880188 y:-12.057999610900879 z:18.14900016784668
- atom: NH2 x: -1.9579999446868896 y:-12.322999954223633 z:20.422000885009766
- residue GLU has 9 atoms
- atom: N x: -9.708999633789062 y:-13.553999900817871 z:19.881999969482422
- atom: CA x: -10.630000114440918 y:-13.32800006866455 z:20.979999542236328
- atom: C x: -9.880999565124512 y:-12.907999992370605 z:22.240999221801758
- atom: O x: -8.704000473022461 y:-13.222000122070312 z:22.416000366210938
- atom: CB x: -11.458000183105469 y:-14.58899974822998 z:21.243000030517578
- atom: CG x: -12.715999603271484 y:-14.678000450134277 z:20.395999908447266
- atom: CD x: -13.70300006866455 y:-13.559000015258789 z:20.702999114990234
- atom: OE1 x: -14.116000175476074 y:-13.442999839782715 z:21.875999450683594
- atom: OE2 x: -14.069000244140625 y:-12.795999526977539 z:19.7810001373291
- residue PRO has 7 atoms
- atom: N x: -10.557000160217285 y:-12.173999786376953 z:23.131999969482422
- atom: CA x: -9.939000129699707 y:-11.720000267028809 z:24.371000289916992
- atom: C x: -9.152999877929688 y:-12.814000129699707 z:25.082000732421875
- atom: O x: -8.093999862670898 y:-12.553000450134277 z:25.649999618530273
- atom: CB x: -11.133000373840332 y:-11.241999626159668 z:25.18000030517578
- atom: CG x: -11.991999626159668 y:-10.654999732971191 z:24.128000259399414
- atom: CD x: -11.949000358581543 y:-11.706999778747559 z:23.049999237060547
- residue LYS has 9 atoms
- atom: N x: -9.663999557495117 y:-14.041000366210938 z:25.042999267578125
- atom: CA x: -8.987000465393066 y:-15.14900016784668 z:25.709999084472656
- atom: C x: -7.636000156402588 y:-15.522000312805176 z:25.11199951171875
- atom: O x: -6.769999980926514 y:-16.047000885009766 z:25.81800079345703
- atom: CB x: -9.890999794006348 y:-16.393999099731445 z:25.76099967956543
- atom: CG x: -10.576000213623047 y:-16.767000198364258 z:24.45599937438965
- atom: CD x: -11.342000007629395 y:-18.08099937438965 z:24.591999053955078
- atom: CE x: -12.46399974822998 y:-18.19700050354004 z:23.55500030517578
- atom: NZ x: -11.987000465393066 y:-18.033000946044922 z:22.152999877929688
- residue ASP has 8 atoms
- atom: N x: -7.442999839782715 y:-15.236000061035156 z:23.82699966430664
- atom: CA x: -6.190999984741211 y:-15.576000213623047 z:23.158000946044922
- atom: C x: -5.249000072479248 y:-14.38599967956543 z:23.02199935913086
- atom: O x: -4.289000034332275 y:-14.437000274658203 z:22.250999450683594
- atom: CB x: -6.478000164031982 y:-16.13599967956543 z:21.766000747680664
- atom: CG x: -7.806000232696533 y:-16.85700035095215 z:21.69099998474121
- atom: OD1 x: -7.918000221252441 y:-17.975000381469727 z:22.240999221801758
- atom: OD2 x: -8.744000434875488 y:-16.29400062561035 z:21.08300018310547
- residue ARG has 11 atoms
- atom: N x: -5.5269999504089355 y:-13.3100004196167 z:23.753000259399414
- atom: CA x: -4.684999942779541 y:-12.119999885559082 z:23.701000213623047
- atom: C x: -3.753000020980835 y:-12.059000015258789 z:24.905000686645508
- atom: O x: -2.9630000591278076 y:-11.131999969482422 z:25.038000106811523
- atom: CB x: -5.535999774932861 y:-10.854000091552734 z:23.65399932861328
- atom: CG x: -6.466000080108643 y:-10.753000259399414 z:22.45599937438965
- atom: CD x: -7.349999904632568 y:-9.517000198364258 z:22.576000213623047
- atom: NE x: -8.53499984741211 y:-9.595999717712402 z:21.73699951171875
- atom: CZ x: -9.552000045776367 y:-8.748000144958496 z:21.80900001525879
- atom: NH1 x: -9.529999732971191 y:-7.751999855041504 z:22.68600082397461
- atom: NH2 x: -10.593000411987305 y:-8.897000312805176 z:21.003000259399414
- residue SER has 6 atoms
- atom: N x: -3.8580000400543213 y:-13.041000366210938 z:25.791000366210938
- atom: CA x: -2.989000082015991 y:-13.07699966430664 z:26.95599937438965
- atom: C x: -1.5839999914169312 y:-13.416000366210938 z:26.474000930786133
- atom: O x: -1.284999966621399 y:-14.574000358581543 z:26.16699981689453
- atom: CB x: -3.4690001010894775 y:-14.126999855041504 z:27.961000442504883
- atom: OG x: -4.433000087738037 y:-13.57800006866455 z:28.847000122070312
- residue GLU has 10 atoms
- atom: N x: -0.7360000014305115 y:-12.394000053405762 z:26.395999908447266
- atom: CA x: 0.6309999823570251 y:-12.565999984741211 z:25.93899917602539
- atom: C x: 1.6269999742507935 y:-11.968999862670898 z:26.92799949645996
- atom: O x: 2.7279999256134033 y:-12.541000366210938 z:27.077999114990234
- atom: CB x: 0.7960000038146973 y:-11.9399995803833 z:24.552000045776367
- atom: CG x: -0.11400000005960464 y:-12.574999809265137 z:23.51099967956543
- atom: CD x: 0.17499999701976776 y:-12.116999626159668 z:22.09000015258789
- atom: OE1 x: 0.09000000357627869 y:-10.902999877929688 z:21.815000534057617
- atom: OE2 x: 0.4819999933242798 y:-12.97700023651123 z:21.23900032043457
- atom: OXT x: 1.3029999732971191 y:-10.935999870300293 z:27.54400062561035
- residue MN has 1 atoms
- atom: MN x: -6.214000225067139 y:-5.52400016784668 z:35.61399841308594
- residue MN has 1 atoms
- atom: MN x: -6.142000198364258 y:-2.124000072479248 z:36.37300109863281
- residue MN has 1 atoms
- atom: MN x: 13.84000015258789 y:-1.5140000581741333 z:61.231998443603516
- residue MN has 1 atoms
- atom: MN x: 8.105999946594238 y:-25.408000946044922 z:15.770000457763672
- residue NA has 1 atoms
- atom: NA x: 4.251999855041504 y:-0.11299999803304672 z:43.87799835205078
- residue NA has 1 atoms
- atom: NA x: 16.698999404907227 y:-13.654000282287598 z:18.052000045776367
- residue F2A has 30 atoms
- atom: PA x: -3.8420000076293945 y:-3.880000114440918 z:34.38800048828125
- atom: O1A x: -5.063000202178955 y:-3.622999906539917 z:35.20000076293945
- atom: O2A x: -2.5329999923706055 y:-3.359999895095825 z:34.85300064086914
- atom: C3A x: -3.6730000972747803 y:-5.749000072479248 z:34.435001373291016
- atom: O5' x: -4.066999912261963 y:-3.2890000343322754 z:32.92499923706055
- atom: PB x: -4.76800012588501 y:-7.072999954223633 z:33.6879997253418
- atom: O1B x: -4.315000057220459 y:-7.23199987411499 z:32.28099822998047
- atom: O2B x: -6.175000190734863 y:-6.676000118255615 z:33.970001220703125
- atom: O3B x: -4.465000152587891 y:-8.342000007629395 z:34.42300033569336
- atom: PG x: -4.432000160217285 y:-8.357000350952148 z:35.915000915527344
- atom: O1G x: -2.990999937057495 y:-8.444000244140625 z:36.2599983215332
- atom: O2G x: -5.375 y:-9.385000228881836 z:36.42900085449219
- atom: O3G x: -4.946000099182129 y:-7.052999973297119 z:36.409000396728516
- atom: C5' x: -5.205999851226807 y:-3.6419999599456787 z:32.145999908447266
- atom: C4' x: -4.960000038146973 y:-3.3380000591278076 z:30.687000274658203
- atom: O4' x: -4.2230000495910645 y:-2.0969998836517334 z:30.54800033569336
- atom: C1' x: -3.4019999504089355 y:-2.1610000133514404 z:29.38800048828125
- atom: N9 x: -2.00600004196167 y:-1.9320000410079956 z:29.775999069213867
- atom: C4 x: -1.0019999742507935 y:-1.4759999513626099 z:28.95199966430664
- atom: N3 x: -1.1009999513626099 y:-1.1349999904632568 z:27.658000946044922
- atom: C2 x: 0.08100000023841858 y:-0.7540000081062317 z:27.180999755859375
- atom: N1 x: 1.2619999647140503 y:-0.6800000071525574 z:27.80299949645996
- atom: C6 x: 1.3289999961853027 y:-1.0219999551773071 z:29.106000900268555
- atom: N6 x: 2.506999969482422 y:-0.9359999895095825 z:29.729999542236328
- atom: C5 x: 0.14499999582767487 y:-1.4479999542236328 z:29.731000900268555
- atom: N7 x: -0.12700000405311584 y:-1.8680000305175781 z:31.024999618530273
- atom: C8 x: -1.4129999876022339 y:-2.138000011444092 z:31.003999710083008
- atom: C2' x: -3.621999979019165 y:-3.5390000343322754 z:28.760000228881836
- atom: C3' x: -4.086999893188477 y:-4.3429999351501465 z:29.964000701904297
- atom: O3' x: -4.868000030517578 y:-5.460000038146973 z:29.570999145507812
- residue HOH has 1 atoms
- atom: O x: -2.4089999198913574 y:8.484999656677246 z:43.20899963378906
- residue HOH has 1 atoms
- atom: O x: -0.14300000667572021 y:-1.7710000276565552 z:48.332000732421875
- residue HOH has 1 atoms
- atom: O x: -16.437999725341797 y:-4.840000152587891 z:18.80699920654297
- residue HOH has 1 atoms
- atom: O x: 16.079999923706055 y:1.4079999923706055 z:30.270999908447266
- residue HOH has 1 atoms
- atom: O x: 21.016000747680664 y:-1.5190000534057617 z:33.81999969482422
- residue HOH has 1 atoms
- atom: O x: -6.730999946594238 y:0.6259999871253967 z:20.711999893188477
- residue HOH has 1 atoms
- atom: O x: 20.124000549316406 y:-7.186999797821045 z:32.66400146484375
- residue HOH has 1 atoms
- atom: O x: -12.125 y:5.327000141143799 z:17.954999923706055
- residue HOH has 1 atoms
- atom: O x: -13.345999717712402 y:10.083999633789062 z:45.89799880981445
- residue HOH has 1 atoms
- atom: O x: -8.428000450134277 y:6.59499979019165 z:30.35300064086914
- residue HOH has 1 atoms
- atom: O x: -18.88599967956543 y:7.940000057220459 z:27.29599952697754
- residue HOH has 1 atoms
- atom: O x: -12.711000442504883 y:-14.880000114440918 z:25.305999755859375
- residue HOH has 1 atoms
- atom: O x: 13.637999534606934 y:-8.12399959564209 z:24.416000366210938
- residue HOH has 1 atoms
- atom: O x: -4.113999843597412 y:-2.135999917984009 z:36.98699951171875
- residue HOH has 1 atoms
- atom: O x: 12.25 y:-8.463000297546387 z:47.82099914550781
- residue HOH has 1 atoms
- atom: O x: 13.711000442504883 y:2.4779999256134033 z:60.03499984741211
- residue HOH has 1 atoms
- atom: O x: 13.440999984741211 y:-24.489999771118164 z:30.077999114990234
- residue HOH has 1 atoms
- atom: O x: -1.406000018119812 y:-12.001999855041504 z:50.17599868774414
- residue HOH has 1 atoms
- atom: O x: -18.986000061035156 y:-14.868000030517578 z:32.553001403808594
- residue HOH has 1 atoms
- atom: O x: -10.734000205993652 y:-14.918000221252441 z:48.27399826049805
- residue HOH has 1 atoms
- atom: O x: 11.635000228881836 y:-24.492000579833984 z:31.9689998626709
- residue HOH has 1 atoms
- atom: O x: 3.058000087738037 y:-2.069999933242798 z:43.28900146484375
- residue HOH has 1 atoms
- atom: O x: 1.7419999837875366 y:-11.512999534606934 z:49.70600128173828
- residue HOH has 1 atoms
- atom: O x: -25.611000061035156 y:13.20199966430664 z:33.939998626708984
- residue HOH has 1 atoms
- atom: O x: -29.035999298095703 y:5.309000015258789 z:24.209999084472656
- residue HOH has 1 atoms
- atom: O x: 0.3100000023841858 y:-5.184999942779541 z:48.196998596191406
- residue HOH has 1 atoms
- atom: O x: -13.279000282287598 y:3.0230000019073486 z:18.819000244140625
- residue HOH has 1 atoms
- atom: O x: -23.634000778198242 y:-16.91900062561035 z:58.840999603271484
- residue HOH has 1 atoms
- atom: O x: -2.9119999408721924 y:-6.1479997634887695 z:54.277000427246094
- residue HOH has 1 atoms
- atom: O x: -1.1610000133514404 y:10.043000221252441 z:47.38800048828125
- residue HOH has 1 atoms
- atom: O x: 29.593000411987305 y:-14.854999542236328 z:40.95399856567383
- residue HOH has 1 atoms
- atom: O x: -12.48900032043457 y:5.578999996185303 z:53.430999755859375
- residue HOH has 1 atoms
- atom: O x: 12.057000160217285 y:2.822999954223633 z:36.19300079345703
- residue HOH has 1 atoms
- atom: O x: 27.393999099731445 y:-13.095000267028809 z:37.10499954223633
- residue HOH has 1 atoms
- atom: O x: -28.520000457763672 y:9.92199993133545 z:38.66999816894531
- residue HOH has 1 atoms
- atom: O x: -5.872000217437744 y:10.038999557495117 z:19.493999481201172
- residue HOH has 1 atoms
- atom: O x: 25.288000106811523 y:-19.02199935913086 z:24.20199966430664
- residue HOH has 1 atoms
- atom: O x: -3.128999948501587 y:-4.6519999504089355 z:39.78499984741211
- residue HOH has 1 atoms
- atom: O x: -15.873000144958496 y:-9.145000457763672 z:20.726999282836914
- residue HOH has 1 atoms
- atom: O x: -5.986999988555908 y:-5.311999797821045 z:55.07600021362305
- residue HOH has 1 atoms
- atom: O x: -22.631999969482422 y:-11.35200023651123 z:32.91999816894531
- residue HOH has 1 atoms
- atom: O x: 15.930000305175781 y:-11.468000411987305 z:48.625
- residue HOH has 1 atoms
- atom: O x: 10.845999717712402 y:10.20199966430664 z:45.08700180053711
- residue HOH has 1 atoms
- atom: O x: -1.7580000162124634 y:-6.790999889373779 z:39.04999923706055
- residue HOH has 1 atoms
- atom: O x: 0.8429999947547913 y:1.375 z:59.32500076293945
- residue HOH has 1 atoms
- atom: O x: -23.67099952697754 y:9.42199993133545 z:51.630001068115234
- residue HOH has 1 atoms
- atom: O x: -7.863999843597412 y:-7.434000015258789 z:35.81700134277344
- residue HOH has 1 atoms
- atom: O x: -14.73900032043457 y:-2.5480000972747803 z:57.404998779296875
- residue HOH has 1 atoms
- atom: O x: -12.61299991607666 y:-3.4010000228881836 z:56.3650016784668
- residue HOH has 1 atoms
- atom: O x: -12.157999992370605 y:14.347999572753906 z:9.017999649047852
- residue HOH has 1 atoms
- atom: O x: -10.178999900817871 y:16.46299934387207 z:7.783999919891357
- residue HOH has 1 atoms
- atom: O x: -26.187999725341797 y:1.3630000352859497 z:32.53200149536133
- residue HOH has 1 atoms
- atom: O x: -19.97100067138672 y:13.097999572753906 z:46.61899948120117
- residue HOH has 1 atoms
- atom: O x: 24.52400016784668 y:-18.415000915527344 z:41.80699920654297
- residue HOH has 1 atoms
- atom: O x: -18.277999877929688 y:-7.002999782562256 z:31.499000549316406
- residue HOH has 1 atoms
- atom: O x: -17.966999053955078 y:-11.64799976348877 z:31.121000289916992
- residue HOH has 1 atoms
- atom: O x: 19.69499969482422 y:-9.303000450134277 z:57.77000045776367
- residue HOH has 1 atoms
- atom: O x: 20.47800064086914 y:-7.195000171661377 z:55.983001708984375
- residue HOH has 1 atoms
- atom: O x: -20.400999069213867 y:-4.622000217437744 z:26.72800064086914
- residue HOH has 1 atoms
- atom: O x: -23.23699951171875 y:-3.7679998874664307 z:26.531999588012695
- residue HOH has 1 atoms
- atom: O x: -21.20199966430664 y:0.3619999885559082 z:21.589000701904297
- residue HOH has 1 atoms
- atom: O x: -1.5870000123977661 y:-0.9639999866485596 z:42.207000732421875
- residue HOH has 1 atoms
- atom: O x: -24.679000854492188 y:-1.2269999980926514 z:32.99100112915039
- residue HOH has 1 atoms
- atom: O x: -27.02199935913086 y:10.708000183105469 z:33.94599914550781
- residue HOH has 1 atoms
- atom: O x: 11.821000099182129 y:-2.0350000858306885 z:62.28200149536133
- residue HOH has 1 atoms
- atom: O x: 13.04800033569336 y:-3.318000078201294 z:60.446998596191406
- residue HOH has 1 atoms
- atom: O x: -16.23699951171875 y:-0.24300000071525574 z:10.319000244140625
- residue HOH has 1 atoms
- atom: O x: -18.378000259399414 y:4.208000183105469 z:55.77299880981445
- residue HOH has 1 atoms
- atom: O x: -17.55299949645996 y:3.2249999046325684 z:51.25199890136719
- residue HOH has 1 atoms
- atom: O x: 9.491000175476074 y:-6.734000205993652 z:43.38999938964844
- residue HOH has 1 atoms
- atom: O x: 23.27199935913086 y:-28.881000518798828 z:39.137001037597656
- residue HOH has 1 atoms
- atom: O x: -13.192999839782715 y:17.964000701904297 z:19.21299934387207
- residue HOH has 1 atoms
- atom: O x: 21.905000686645508 y:-4.89900016784668 z:35.422000885009766
- residue HOH has 1 atoms
- atom: O x: 2.9800000190734863 y:-5.75 z:44.49599838256836
- residue HOH has 1 atoms
- atom: O x: 4.548999786376953 y:-9.392000198364258 z:42.6619987487793
- residue HOH has 1 atoms
- atom: O x: 4.017000198364258 y:-12.059000015258789 z:46.9379997253418
- residue HOH has 1 atoms
- atom: O x: 6.761000156402588 y:-12.234000205993652 z:47.72800064086914
- residue HOH has 1 atoms
- atom: O x: 5.676000118255615 y:-16.034000396728516 z:45.32099914550781
- residue HOH has 1 atoms
- atom: O x: -7.611999988555908 y:-15.508000373840332 z:46.83700180053711
- residue HOH has 1 atoms
- atom: O x: 10.524999618530273 y:-10.456999778747559 z:40.52899932861328
- residue HOH has 1 atoms
- atom: O x: 6.034999847412109 y:-0.2840000092983246 z:30.01300048828125
- residue HOH has 1 atoms
- atom: O x: 7.501999855041504 y:-17.215999603271484 z:20.152000427246094
- residue HOH has 1 atoms
- atom: O x: 7.015999794006348 y:-19.85099983215332 z:17.801000595092773
- residue HOH has 1 atoms
- atom: O x: 10.140999794006348 y:-20.489999771118164 z:16.503999710083008
- residue HOH has 1 atoms
- atom: O x: -28.062999725341797 y:-9.74899959564209 z:39.11600112915039
- residue HOH has 1 atoms
- atom: O x: -13.135000228881836 y:6.835000038146973 z:48.64500045776367
- residue HOH has 1 atoms
- atom: O x: -12.461000442504883 y:13.706000328063965 z:21.788999557495117
- residue HOH has 1 atoms
- atom: O x: -21.52199935913086 y:-11.213000297546387 z:47.9640007019043
- residue HOH has 1 atoms
- atom: O x: -22.802000045776367 y:-13.795999526977539 z:49.27899932861328
- residue HOH has 1 atoms
- atom: O x: 17.39699935913086 y:-18.308000564575195 z:38.902000427246094
- residue HOH has 1 atoms
- atom: O x: 13.23799991607666 y:-16.785999298095703 z:38.619998931884766
- residue HOH has 1 atoms
- atom: O x: 11.210000038146973 y:-8.220000267028809 z:18.395999908447266
- residue HOH has 1 atoms
- atom: O x: -1.9579999446868896 y:10.479000091552734 z:54.09700012207031
- residue HOH has 1 atoms
- atom: O x: 18.85700035095215 y:-12.720999717712402 z:17.263999938964844
Once we have the coordinates of an atom we can compute distances between atoms or angles. We can also align two structures rototranslating the two to minimize their distance. We will not cover this and many other features that are provided by Biopython, such as Pyhlogentic analysis tools, interface towards pathways in KEGG, clustering, etc. If you are interested can find all the features available in the Biopython tutorial.
Exercises¶
- Write a python script that retrieves all the information present in SRA regarding PacBio sequencing performed on E.coli strain K12 (query term is “E.coli K12 wgs PacBio”). Print the number of results and for each id report the title, the accession id, the total number of spots and total number of bases sequenced.
Sample output:
Entries found: 10
Results for id: 6705337
- acc="SRR8154667"
- total_spots="163482"
- total_bases="1561717136"
Results for id: 6705336
- acc="SRR8154668"
- total_spots="163482"
- total_bases="897324802"
Results for id: 6705335
- acc="SRR8154669"
- total_spots="163482"
- total_bases="1570240924"
...
...
Show/Hide Solution
- Write a python function that reads all the entries of a blast alignment file in .xml format (like blast_res_apple.xml and outputs all the HSPs (see example below) having bitscore > B, alignment length > A and minimum percentage of identity > I, where B, A and I are input thresholds. Hint: implement a filtering function: filterHSPs(align, minBitscore = 0, minAlignLen = 0, minPercIdent = 0.1).
Alignments of MDC020656.85
MDC020656.85: 1939-2593
gi|125995253|dbj|AB270792.1|: 201263-201917
Score:820.917 AlignLen:579 Id/Len:0.8812785388127854
MDC020656.85: 1446-1935
gi|125995253|dbj|AB270792.1|: 306490-306017
Score:582.873 AlignLen:428 Id/Len:0.8629032258064516
....
....
that is reporting the HSP with query start-end position, subject start-end position, score, alignment length and number of identities / alignment length.
Show/Hide Solution
- Write a python function
retrieve_sequences(search_term, number, outfile)that retrieves the firstnumberof sequences from NCBI’s “nucleotide” database having a search termterm(hint: use term and retmax parameters of Entrez.esearch) and stores them in a fasta fileoutfile(hint: use SeqIO.write). Test your code retrieving the first 5 entries having search term “starch AND Malus Domestica [Organism]”
Show/Hide Solution
- Write a python function that aligns the sequences in the file created
in exercise 3. (here you
can find mine) against the NCBI nr database limiting the hits to the
Malus Domestica organism (parameter entrez_query=‘“Malus Domestica”
[Organism]’ in qblast)and prints to screen the following info for
each hsp:
- The title;
- Score and e-value;
- The number of alignments on the same subject, the number of identities and positives and the alignment length;
- The number of mismatches and the list of their positions (hint: you can use the match string and look for ” “).
Show/Hide Solution
- Write a python function
getPublicationInfo(title_term,other_term)that retrieves the first 20 pubmed publications having thetitle_termin the title andother_termsomewhere else in the text (hint use: “Title” and “[Other Term]” as esearch parameter term). For each publication print:- the title
- authors
- journal
- year of publication (hint: get and split properly the “PubDate” entry)
- a link to the pubmed entry (hint: it is the string “https://www.ncbi.nlm.nih.gov/pubmed/” followed by the pubmed id (“eid” entry of the dictionary “ArticleIds”). es: https://www.ncbi.nlm.nih.gov/pubmed/26919684
Hint: to see how to combine search terms test them here: https://www.ncbi.nlm.nih.gov/pubmed/advanced.
Test your code calling getPublicationInfo("apple","drought")
Show/Hide Solution
- Write some python code to retrieve the structure of two forms of the
aspartate transcarbamoylase (PDB ids: 4FYW and 1D09). If you are
interested, read more about the Aspartate Transcarbamoylase
here. Write a function that
gets the .cif file name and prints:
- the number of chains, residues and atoms present in the file;
- a histogram of the residues (plotting it with matplotlib) that are not water (encoded as “HOH”);
- a link to an online tool to visualize the 3D structure. The link will be “http://www.rcsb.org/pdb/ngl/ngl.do?pdbid=” followed by the PDB id of the protein (e.g. 1d09).
Show/Hide Solution